













| General Information: |
|
| Name(s) found: |
CE02527
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 770 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
lipid storage
[IMP]
reproduction [IMP] |
| Molecular Function: |
zinc ion binding
[IEA protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFIADCDRTC EKPADAFLDA NASLLPSLSR FSLVSGNSFI KTMGDSGVEP VIPSPSTSSN 60
61 NPVEELQCTI CLSTRFSQEC RIEGCNHSFC FSCISEWVCQ SLRPSCPMCR KDVDKVSYDF 120
121 SEQKLGQKEI AIKEYRSGHM SLTPTDRVQL LSERRLVVRN LMHSYKVVGL LSSEISGIEE 180
181 SKEPDEILKN ALIELAAIVT QHIQKAEISL SDLRTEQARI NKSIVFNTVT FRRMVYTSKV 240
241 KVMYPDLKKV KIKPSDIARE PDRFRNVITN FLIAEFDAIP FQNQPKLAGH KWRTKFLGSC 300
301 IDMQQKQRYA AQIYELMLSR PSGDHLFRRE LNEILTPVSA VHIQFLDDHL DAIIAIEKLT 360
361 IEEFYNEVYY DSIYNSFSSN FFDYNSMGRD DPQSRFEALF NQIHRDIGST NDWMDTYVAG 420
421 RLLRQSSNTD GTDRATNNDD DSDRSESPEE RVPTPMPDED PREQSLARAY YEQMLEMPNR 480
481 VPLSMRAYHM MRDRDVRVRY PTTEEPVPVE PVTFGQTTRT LPISQQPTTS RSHMTLRSAM 540
541 RRLDSDRPTD EEIPPIESRP QTRREIERDR LLQRVREFDN RSRGLSSLFR HPASSDYTSR 600
601 RFTSDGYSGL VMLSPRLNST TTSSTSIPVY QPRTMTSNDF AHLNRTRSIL ERRHFPQPPR 660
661 EQRIEEVEQL FNRAYQNMQQ THQAIQEHAS TSNAPTQIPR PATPAPAISD EESRSLAEIV 720
721 ATINQEDADN GLDFISVAHR TRSARRAAEN GHPPEGPPAK RSTNSRRDRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
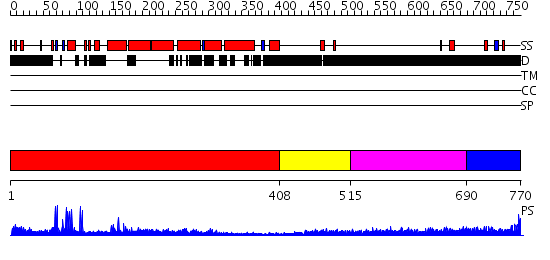
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..407] | 8.69897 | Heavy meromyosin subfragment | |
| 2 | View Details | [408..514] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [515..689] | 4.204997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [690..770] | 4.189993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)