













| General Information: |
|
| Name(s) found: |
nduf-2.2 /
CE00702
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 474 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
electron transport
[IEA nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP growth [IMP |
| Molecular Function: |
NADH dehydrogenase (quinone) activity
[IEA NAD or NADH binding [IEA metal cluster binding [IEA electron carrier activity [IEA oxidoreductase activity, acting on NADH or NADPH [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSRSLHPLR AVACARPAIS NRDSHTIWYP DAKFERQFKT GGTLGKLWMS ERVSDFDDQI 60
61 GLDKLEKLAY SDPVLSDNYE GKKREKNLEN MILNFGPQHP AAHGVLRLVL KLEGEVIIKA 120
121 IPHIGLLHRA TEKLIEHKTY TQALPYFDRL DYVSMMCNEQ AFSLAIEKLL GIDVPPRAKY 180
181 IRILFGELTR IQNHIMGITT HALDVGAMTP FFWMFEEREK LFEFSERVSG ARMHANYVRP 240
241 GGVAWDLPVG LMDDIYDWAV KFPARIDELE DMLTENRIWK ARTVDIGLVS ASDALNWGFS 300
301 GVMVRGSGIK QDVRKTEPYD AYADMEFDVP IGTKGDCYDR YLCRVEEMRQ SLNIVHQCLN 360
361 KMPTGEIKSD DHKVVPPKRA EMKENMESLI HHFKFFTEGF QVPPGATYVP IEAPKGEFGV 420
421 YLVADGTGKP YRCFIRAPGF AHLAAIHDVC YMSLIADIVA VIGTMDIVFG EVDR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
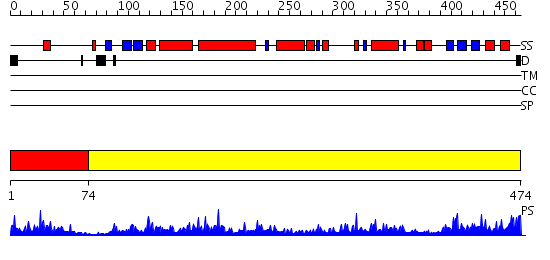
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | 15.30103 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus | |
| 2 | View Details | [74..474] | 122.0 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)