













| General Information: |
|
| Name(s) found: |
eif-3.D /
CE00291
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 570 amino acids |
Gene Ontology: |
|
| Cellular Component: |
eukaryotic translation initiation factor 3 complex
[ISS |
| Biological Process: |
locomotory behavior
[IMP hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP translational initiation [IEA reproduction [IMP |
| Molecular Function: |
translation initiation factor activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALPKFELLS LADNTVGWGP LASSSSADEP VPFQQFNKAD RIGRVADWIG VDRFYRRGNE 60
61 RYNERVYGSA ANAGSQFDYI HGMDEHNFQL VDTSKPMARN PQRNFRVRQM HLRKMMQKEN 120
121 EKREMVNQST NLRMKRSIAK EQQRAFKMWQ RRGGNARQGQ RGQGGRFGGD RPKERLPSVQ 180
181 VRPEWVVLEE MNLSAFSKLA LPNIPGGDDI GDHQYGSLQY YDKTIDRVSV KNSIPLQRCA 240
241 GVFYNVTTTE DPVIQELAQG GAGNVFGTDI ILATLMTAPR SVYSWDIVAY RVGDKLFFDK 300
301 RNTRDILNPV ETLTVSETSA EPPSFDGNGI NNAKDLATEA FYINQNFRRQ VVKRNDAGFT 360
361 FKNARAPFED EETGESGTAY KYRKWNLGNG VDGKPVELVC RTELDGVIHG LGNETQTLTI 420
421 KAFNEWDSTQ SGGVDWRTKL DVQKGAVMAT EIKNNSAKVA KWTLQALLAG SDTMKLGYVS 480
481 RNNARSTQNH SILLTQYVKP TEFASNIALN MDNCWGILRC VIDSCMKQKP GKYLLMKDPQ 540
541 SPVIRLYSLP EGTFESERES SDEENSDDDQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
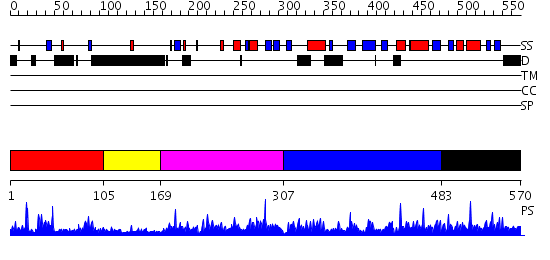
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..104] | 1.029991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [105..168] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [169..306] | 1.032955 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [307..482] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [483..570] | 2.030965 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)