













| General Information: |
|
| Name(s) found: |
SPCC1020.13c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 669 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
cellular lipid catabolic process
[IC]
|
| Molecular Function: |
metal ion binding
[IEA]
phospholipase activity [NAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQESSQEEPV HYVQWFYLSN YDLYDETSSM ELTGAQSWRR FLNYDNNALE AKYNEIITEE 60
61 VSQEPNQKNV IVGISGLHVV NLPTLTLKPL YWPSASKNDT LRVVRGLWMY EDTGLPVINE 120
121 LSDMLEEGYQ EVKPYAVNWD NVLKEEEEGK DSQDDPSVLS HKRNKSSKEL IDAMQWPLKP 180
181 TSSGKYVFYE DGYRALICQG GLLSYFSKGT QSRRTSIKGM PVVRDFPYFE DDGFNGPKQV 240
241 TDLFLVVHGI GQKRSETEER FLFTKTCNVF RSLIQIQKNI MKEDPLIRND YEPQLLPICW 300
301 RNKLNFNSYI KPVAGDEGRV EDEEFEENRF SIEDIEIDSI PAVRRLLGDV MSDIPYYMSH 360
361 HKESIIKSVI REANRVYHLW KDCNPYFLEN KGRIFIIGHS LGSTVVFDIL SLQPTFVKEI 420
421 TIDDDESCFI FDTNGFFCFG GPVGFFLHLN QQSIIPRRGR GSSRYEINKN FINSDVPKSY 480
481 TYDGYDRYGC LAVDSFYNIY NHLDPVAMRL NPTVDLSFSK GIHPTRILFT RKSSSILRMI 540
541 SHSNTDPVEL RSYNQSLQQN KNNEPTAVVP LEPEQTVELE TRNFRREERA KWRMQELNEN 600
601 SQLDYVFTSA HGVISNKYLS MLSAHSSYWS SEDLACFLVV ETGRNFGIAN SIEQFRGKHM 660
661 PRIFKGDSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
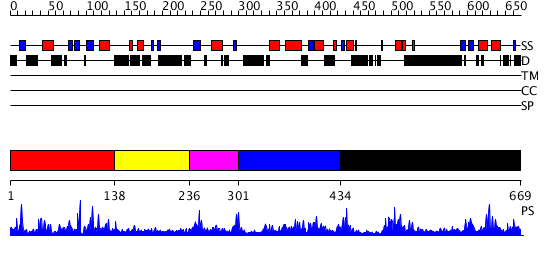
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] | 3.054987 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [138..235] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [236..300] | 2.084992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [301..433] | 1.087987 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [434..669] | 76.744727 | No description for PF02862.8 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)