













| General Information: |
|
| Name(s) found: |
lsd1 /
SPBC146.09c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1000 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Lsd1/2 complex [IDA nuclear chromatin [IC |
| Biological Process: |
chromatin remodeling
[ISS cellular response to heat [IEP positive regulation of transcription, DNA-dependent [IMP regulation of meiosis [IMP histone H3-K9 demethylation [IMP antisense RNA transcription [IMP |
| Molecular Function: |
DNA binding
[IEA]
histone demethylase activity (H3-monomethyl-K4 specific) [NAS] nucleosome binding [IDA histone demethylase activity (H3-K9 specific) [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMDLSSKDAL SDVLKDTHAQ SSDPSLWAIF GHQSSDNVHE GGNESVSVEQ EIFDLSQLSE 60
61 RPVSETVSKA SIPTNINGLS QVKPSALEKS QEVLSLQKLP IKGRRPAGRR GRPALNTSNS 120
121 LERNGTRYVS AEAPISVKSS IPAIPRVTFE RLCYESAIAS NLPPNALSPL EAEMLSEILE 180
181 NPTWLSLYLS IRNGICYLWH RNPTLYVSFN EALGIVREKK AFPLASLAFE FLSRNGHINY 240
241 GCIYIISSLK LDESLSQKTV AIIGAGMAGI SCARQLTNLF AQYEQDFLSR GEKPPRIVIY 300
301 EASERLGGHI YTHMVPLSDN EVSEKSSLAT TVNATNECMV NLLTDSLIGM PTLDSDPLYI 360
361 ISSQQLSLDA VHTRNREFIL HDIENGRIDT EHVQRIFRLF DALLFYFNAS ASKQPLHSLI 420
421 TPPEQEFIQK LDQIGWYISI EAFPLQIKDT LSEFLGNSAN TLTSLLHLTV LDLKIFEWFK 480
481 EYLSQSLSVS LENVYPGSIP NLNLLLGENV ASYSFKHGMA DMLNSLASTP SPLPILFDQC 540
541 VHTVKLEDNT VNLSFVNETT VSVDKVVICI PMDKLNTHLI TFEPPLEEKK LKAIDRCHFT 600
601 NVKKVILIFK TQFWEPNISI FGSLPQDSGR NFIFNDCTRF YEHPTLSVFV KVEGIDFMKD 660
661 DDIVNGIVSQ LKKVYKPKSE AINPIRTIIS NWENNSYTNH SSYQISNLFL EEDYAILSEP 720
721 IDNTVFFASE AISQKNSGSI RGAFDSGILA ARDVLASLIG NVVLPNTLVI EENLEQPRKT 780
781 YGTKRNAQQA LGKEGERENK EKRISYHTEY LRLRQKRLDK EQQECDLLIA ELLGPSPVPP 840
841 SRPSANPYLL YQKTQWHVCK TLADQDKQRV TGDPEARATK NEIRAKLGKT WRALDSLGKQ 900
901 PWVDEINARR ANYSTRLEEY QRQINSYNVR VAQIKSEHQR RCESQPIPED EAKLKLLAEQ 960
961 EDEHLHPEKE GMSVENSDDD YHDDLDYEDS ISEVFPDNFS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
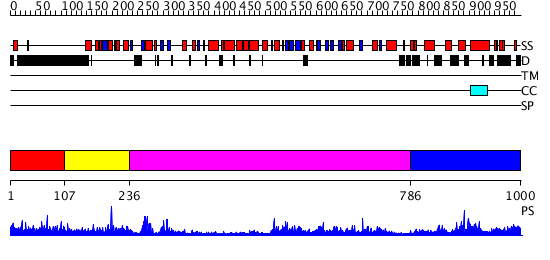
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 1.065995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [107..235] | 26.161151 | No description for PF04433.4 was found. No confident structure predictions are available. | |
| 3 | View Details | [236..785] | 74.0 | The structure basis of specific recognitions for substrates and inhibitors of rat monoamine oxidase A | |
| 4 | View Details | [786..1000] | 4.045757 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)