













| General Information: |
|
| Name(s) found: |
SPBC13G1.02
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 414 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
GDP-mannose biosynthetic process
[IC |
| Molecular Function: |
GTP binding
[IEA]
mannose-1-phosphate guanylyltransferase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISSAVILVG GPSRGTRFRP LSFDVPKPLF KIGGREMIYH HLAALSKIES VKDVFLVGFY 60
61 DESVFKDFIN EVASHFPSFN RIKYLREYNC LGTGGGLYHF RDQILKGHTS NVFVMHADVC 120
121 CSFPLQELLN VHHEKKALVT LMATKVSKED ASNFGCLVEE PSTGRVLHYV DKPSSYLSNI 180
181 ISCGIYIFDA SIFDEIKKAY ERRLEEVEKQ LRSLDEGMED YLSLETDVLA PLCSDSSKAI 240
241 YAYNTPEFWR QIKTAGSAVP ANSLYLQKAY HDGTLPKPDT EAEIIQPVFI HPNAIVSKGA 300
301 KIGPNVSIGA RVRIEDGARI RNSIIQEDCE ISANAVVLHS ILSRHCKIGK WSRVEGSPTL 360
361 PSQHSTTIMR NSVKVQAITV MGADCIVHDE VRVQNCLVLP HKEIKVGLVG EIVM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
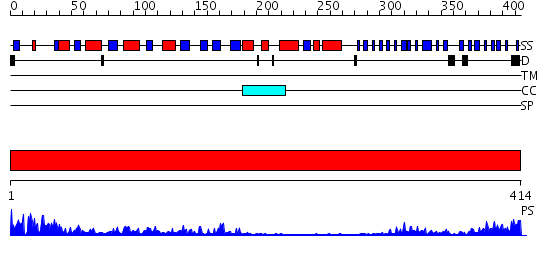
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..414] | 43.045757 | N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain; N-acetylglucosamine 1-phosphate uridyltransferase GlmU, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)