













| General Information: |
|
| Name(s) found: |
mfh2 /
SPAC20H4.04
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 783 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA |
| Biological Process: |
DNA repair
[ISS |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] ATP-dependent 3'-5' DNA helicase activity [IC 3'-5' DNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIVLRDSSDY SDSEVDLPSI EVNGERGNVP TSVLCNVEGS LLNSNPALRQ PNLKCIKHNQ 60
61 DVENTTFQLD LRGCRVPSSQ PQVITELENN IDIPKNIDAM QNWIFPQTQQ YRNYQKEFCE 120
121 QALFHNLLLA LPTGLGKTFI AAVVMLNYFR WFPESKIIFL APTKPLLLQQ RVACSNVAGM 180
181 SPGATAELNG EVSPDRRLFE YNTKRVFFMT PQTLQNDLKE HLLDAKSIIC LIFDEAHRAT 240
241 GNHSYAQVMR AVLRSNSHFR VLGLTATPGS STASVQKVVD CLHISKLIVR NEESIDIRSY 300
301 VFHKKIQLIK VTISSEMNIL KSDFANLYRP YFNFLRQKKL IPINCECLNI KAYTLFVSLR 360
361 KYSFSSKNVQ SKEKSKIMSC FTLLISCAHI TYLLDCHGII QFYQKLVETK NKAEGKGSGQ 420
421 SFWLFTSKPF AFYLEHLHNK IQGLSLNHPK MNHLLELLKE HFKDTSEGYQ NQRVMIFTEF 480
481 RNTAEYITTT LLAIRPMVRA SLFIGQANSA YSTGMNQMQQ KETIDQFRAG VINTLVATSI 540
541 GEEGLDIGDT DMIICYDASS SPIRTIQRMG RTGRKKSGKV FVLLTEDCED SKWERSQVSY 600
601 RRVQKVIESG KKIALKKDVP RLIPSNIQPI FKFQALQNNA DATLILNSYN NNSSSLSPVN 660
661 TLANQAHSRS KRYLPFIVDD VFEDMESNLR VPTEDAKIKR FKSDYRSCIY NARRNVFSKP 720
721 TYMGDKLTKF AKVPHSLLTL SIYRRGRLLQ QCSPSSVTKY LKYEEKFKRK RMKKTSNALF 780
781 QST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
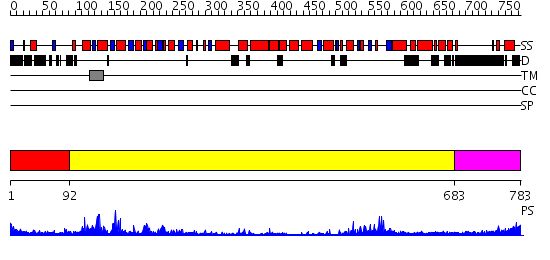
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [92..682] | 45.0 | Crystal structure of Pyrococcus furiosus Hef helicase domain | |
| 3 | View Details | [683..783] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)