













| General Information: |
|
| Name(s) found: |
SPCC74.02c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mRNA cleavage and polyadenylation specificity factor complex [IDA cytosol [IDA |
| Biological Process: |
mRNA cleavage
[IC |
| Molecular Function: |
RNA binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNWNSVRNV SSDRQTSKTS ENPPHTSNEY SGKPEFINLS PDLEENLDEK LMSAFPGLEP 60
61 HVFQHSQSPL SHKDASLLAT MPSVASSNPS LISSGSSQTG SPSQSLSSNK EPSSPGISPS 120
121 NDSQSQNTNH TSISANPYVN NPSHTSRNPD SGSSLNTASH EVPSSKSDVN VQMLARLKSK 180
181 SRQKISSSDP LEDLRLTLTE CLNPINIVQA PKECAAILVN LMSNITQDDQ KLVFLDLLKS 240
241 KVGNSIYSQL VDGGRKLFLP KLRNWFVSAI RSKHDELIHL ILLVLANLPL TTEKLAEVKF 300
301 GKPILIVKKK STNSVIRQLA ENLSELAEKS FTIEQNRENE KSSTKNDSTV SSSAVVMAPA 360
361 GPAMAPSASN KPSASSTTKS SNSKSKKKVT SISGTSFFKN LASSTKPTSA SSSTKAPLTK 420
421 QQTNPSTPLS SIMAGLKGRE KEKDKDSGIS SENVSNNREE LPSFRKRSSS SRQSEEIASL 480
481 QAENAVFSSD PASNDEKTGN KKRKKKSVSW KPDNDLVQVK FIESLNEEGA ASVKTPHIYG 540
541 NARDMDRQEA RVAFGSHVED DVENEIIWYK PVPIKFEISK DEIHPRGYKC GGNERNLTPE 600
601 ATSEIEREKN ESKDISTFNI ILDLPVIREF DDSRPPAHIK LVSSDTQATT ELGFNGLVQQ 660
661 VSENNTNAYS ATSNSQLSSI FSNLSSSISD ASSNVLQNPS LSIPNYSNAI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
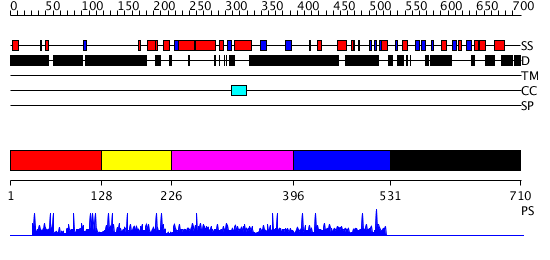
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [128..225] | 1.003975 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [226..395] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [396..530] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [531..710] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)