













| General Information: |
|
| Name(s) found: |
jmj3 /
SPBC83.07
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 752 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC Lid2 complex [IDA cytosol [IDA |
| Biological Process: |
chromatin remodeling
[IC negative regulation of transcription [IC regulation of transcription, DNA-dependent [IC histone demethylation [ISS |
| Molecular Function: |
histone demethylase activity (H3-K36 specific)
[ISS specific transcriptional repressor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQQNVKEKHA GKSDTSVSSL ECEIDYHIEG SDGIPVVEPK ISEFVDMESF IRRVERLGKK 60
61 YGAIKVVRPS SVLNPWNEDT MKPSDVKMDL WLERMVRRKG EYFEIQSDID HGSAGPKKPT 120
121 DMNVDDRFPS NAGSSNDFEN NVAKAIAYYW RSLTHDSLWY GYTNRPSIPF YFIPSISAAQ 180
181 KDRVHLRSNT LINTWPNVGH LFAGKWKTTL PWRVESPELH AVQVHLGGSS LQWYVIPSAH 240
241 SESFKKLAGK LAQDEHWRCS DFLLHQNILF PPSTLVQNGI VTYSTVLKQD ELLITFPGTH 300
301 HSAFCLGDAV LRRFVFRSPR SASNYEFSNL RRLMVSESLY SSKSLWPHSH KPQRACSQKF 360
361 LDEFYLHDLP ESNIHDSGNF HPIHSSVDNN SFSQRDFDSP NSINPPSPLM SNHESASTEH 420
421 FNSTTTTEKE LSSLHVGEER KNRSLPLSLI WNSKAREEYI KKQKEENGDN IEFSHFDPLY 480
481 TRPSSHPLHP PPILGLPVPA QFARGELFLG RILEDRVSEH MLLLECEKSD VVEVPYECIL 540
541 TSSSAAGRRE SSYYNPALKA PNIVYDDGVP INWNEYSELP SLDRFVLPKL LPGKPIEFTP 600
601 PISVEPTSIK TIAAEESSEP TSSVDVAPTP VEDVNVNLES ISNTNESVVD LSDPLVSKNG 660
661 FEDVERSSVA DLEEDVLETR SSIFETSDID DRLTVIDRSQ SVVPSESEFS IAGANLTRRN 720
721 AVDFSVSLDT YELYVSDEVE NVDDFSLFPS LE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
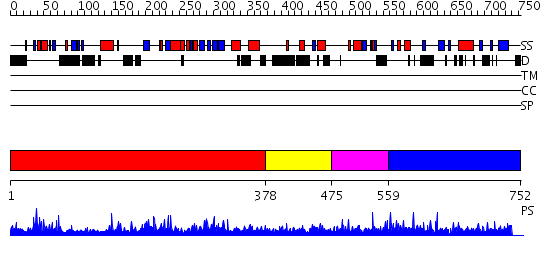
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..377] | 108.0 | No description for 2p5bA was found. | |
| 2 | View Details | [378..474] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [475..558] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [559..752] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)