













| General Information: |
|
| Name(s) found: |
snt2 /
SPAC3H1.12c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1131 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC Lid2 complex [IDA spindle pole body [IDA spindle [IDA |
| Biological Process: |
chromatin remodeling
[IC regulation of transcription, DNA-dependent [IC |
| Molecular Function: |
DNA binding
[IEA]
zinc ion binding [IEA] small conjugating protein ligase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLVIEADSNL LFTAMFDLDK NTNIESNHVK IGNKNTTRRL IIKSSKNSVR IAYAPPEKHF 60
61 VDVTDRFLLP ETETQNLKTR LGIFELEPLP PNGLVCCVLP NGELIQPNDF VLVNSPFPGE 120
121 PFQIARIISF EKSRPCVSTN LYDSVRLNWY FRPRDIQRHL TDTRLLFASM HSDIYNIGSV 180
181 QEKCTVKHRS QIENLDEYKS QAKSYYFDRL FDQNINKVFD VVPVTQVKNA PDDVLEDLFK 240
241 NYDFIVTEYG KGRALLNEPS NCKVCKKWCA FDFSVQCADC KKYYHMDCVV PPLLKKPPHG 300
301 FGWTCATCSF ATQRKKSTFQ KENANVDANH ATENNLEGQA TQKSVSILKG HNKALSNVSL 360
361 QEDHGKRRNL KSLRSSRNLH QQSRKSLDEN KPNSFSNVSK LKRLPWNMRY LDLKSDLTVE 420
421 KKSDIYPSRA RISISPMLPT SSEDNLHPLQ PLTTADEEMD LDLKSDERFK VDIPTFFERW 480
481 PFLKDLPLKG YLFPLCEPNL QSAMLLVPIT YSDALLDDYL CSCWNLWKKL RLPVSAFVFL 540
541 ELTITALYET KLSPAAAFEK LKSWMPGFGD PKNCTGKRVD EHKINSLVKE FGVSLQCFVE 600
601 KLKFEYSLKE IFFSFLSWAS SPKGLNTFKK LSDSSLSTTT TDSHGLPTCC YDIGMYDLQK 660
661 ILKLKKTPIC RWCHSKRSSE WFVAPPIEES SPKDKSKIVA LCQRCGYVWR YYGYPLQQAT 720
721 PSDLRNCDFE PVKKRKADWD HLSNHDNEVK KENNRIRNAS SLMENPRVST KTFDNFTLTH 780
781 DSTINVKADT VKRARQNNIK NKDDVNFSED RKKCCALCGI VGTEGLLVCF KCGTCVHERC 840
841 YVCDDYAENE QMLVSASHLS GRTTRNSASP GIVSGKKSYA KKDQVLSWAC LSCRSNDNLG 900
901 QNNDNHCVLC LQSASHSLMK KTVEGNWVHL ICASWTPDVY VPAEESEPVC GIAQLPPNRW 960
961 EKKCEVCGNS FGVCVSSPNS GLTSHVTCAE KANWYLGFEF VKQDQSPFSM LSNLKSLSFF1020
1021 GNVTEINTNK CMINSWTSLR PVLFGPSEQL PRNFLLRNDI VPNTNNSAWS EYIRNLYPKA1080
1081 YIYLLQYTIA VCKPTIAPTN VACCCSKCNS TMSPFWWPGN ICQACHCLRV E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
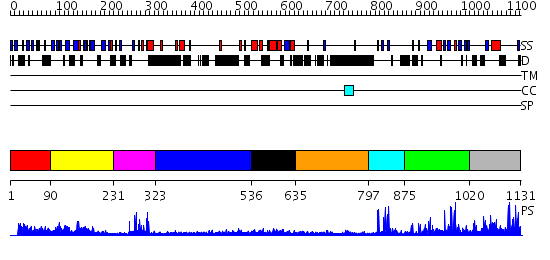
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [90..230] | 18.69897 | Crystal structure of the proximal BAH domain of polybromo | |
| 3 | View Details | [231..322] | 5.522879 | No description for 2ysmA was found. | |
| 4 | View Details | [323..535] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [536..634] | 6.522879 | Solution structure of the myb-like DNA-binding domain of mouse MTA3 protein | |
| 6 | View Details | [635..796] | 3.03 | Erythroid transcription factor GATA-1 | |
| 7 | View Details | [797..874] | 3.522879 | No description for 2puyA was found. | |
| 8 | View Details | [875..1019] | 13.431997 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1020..1131] | 1.205995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)