













| General Information: |
|
| Name(s) found: |
sld3 /
SPAC24H6.06
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 668 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA DNA replication preinitiation complex [IDA nuclear replication fork [IC |
| Biological Process: |
DNA replication initiation
[IGI cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNDHASKKS FCIKAPSNWE KSYLEVWPLV TVPRQCICLR WCISKEYHEF TCSSLQFIVI 60
61 RPAGTSVLLG RVKSSKANQL VGIEHVEGSR YALIFLSEKL DFKSLKVIAN HQLTKSSKSL 120
121 SNVSNKPLGD QLFRSNSLMS PSLLKKELHR IQSDASQANE RESQAPHSFV THDLISSSKD 180
181 GNSLTHEFAN DSVTEMVQDY TPSCSRDVKS LLDHLYNSYF YQLLMTKTPV VFYVKQMVGK 240
241 TRQLAVEVHN HVEEKALVDE LLKFLDNLKS VDDRKSRLLQ CFESHLNYKA WHLEFENEAH 300
301 QYEIKGYRLW LQNILNRENC QITKLDFERE FSQLKLKDLL DSRDSGKRKS RKKNAKTLNP 360
361 FETAQLKLEF TFDGLCIRRT IEQNATERSE DLLLKFCKET IVPYYSSKFP RITRNLLEKC 420
421 NGLDLLPERS HKHRHSAPPR SKLISSKSEA GRALPGNTSG ASISNTSSPH SEASISKDYE 480
481 ILKRRRSNSG VHSLTRSDSS FNGFERDTRR RSSDIARIKN REINLPSSSL SKQRNSMHDI 540
541 STNFPRRNLS FTEKLTMASL QGQSEESVQP KTTSSLSRSK TLSILEGSVS KRSEPSMDSI 600
601 LVQATPRKSS SVITELPDTP IKMNSLDKAS ACTVENHIVT ESPAHKSNKA QLFVCVPTTP 660
661 VKKKSASP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
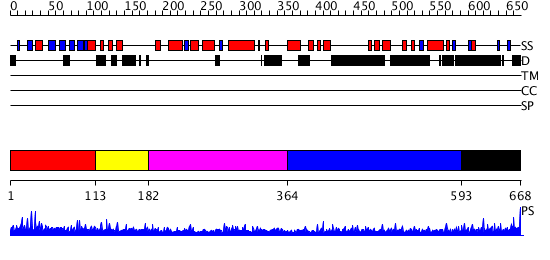
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [113..181] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [182..363] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [364..592] | 4.096996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [593..668] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)