













| General Information: |
|
| Name(s) found: |
top3 /
SPBC16G5.12c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 622 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS mitochondrion [IDA RecQ helicase-Topo III complex [TAS |
| Biological Process: |
nucleolus organization
[IMP DNA-dependent DNA replication [IMP nuclear division [IMP chromosome segregation [IMP resolution of meiotic recombination intermediates [TAS replication fork processing [NAS] DNA topological change [IEA] DNA metabolic process [IGI DNA unwinding involved in replication [IEA] maintenance of rDNA [IMP negative regulation of DNA recombination [TAS |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IPI DNA topoisomerase type I activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRVLCVAEKN SIAKSVASIL GGGHVRRRDT RSKYVKNYDF SFNFGGNVGS SDVTMTSVSG 60
61 HLTEASFPSE YSSWSSVPQD VLFDAQIITS VSKNAEVLAD NIKKEARNAQ YLYIWTDCDR 120
121 EGEHIGVEIS NVARASNPSI QVIRADFNNL ERSHIISAAK RPRDVSKNAA DAVDARIELD 180
181 FRLGAIFTRL QTIQLQKSFD ILQNKIISYG PCQFPTLGFV VDRWQRVEDF VPETYWHLRF 240
241 VDKRQGKTIQ FNWERAKVFD RLTTMIILEN CLECKTAKVV NITQKPKTKY KPLPLSTVEL 300
301 TKLGPKHLRI SAKKTLELAE NLYTNGFVSY PRTETDQFDS SMNLHAIIQK LTGAQEWDSY 360
361 AEGLLAGDYR PPRKGKHNDR AHPPIHPVQM VHRSALPSQD HWKVYELITR RFLACCSDNA 420
421 KGAETLVQVK MEEELFSKKG LLVTEKNYLE VYPYEKWESS DQLPEYRLHE EFQPHILDMM 480
481 DSSTSSPSYI TEPELIALMD ANGIGTDATM AEHIEKVQER EYVIKRKKRG QGVTEFVPSS 540
541 LGVALAKGYD EIGLEWSLTK PFLRKEMEVQ LKNIENGQLN RNVLVHMILT QFRDVFHLTK 600
601 QRFDCLKNSC RVYLMSHNEP QT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
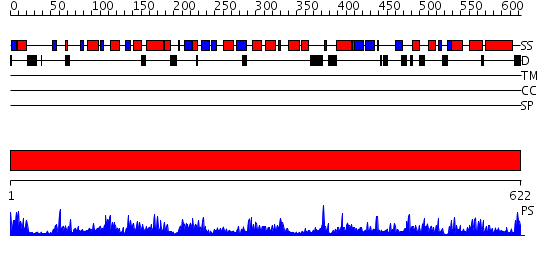
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..622] | 179.0 | DNA topoisomerase I, 67K N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)