













| General Information: |
|
| Name(s) found: |
alp4 /
SPBC365.15
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 784 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA equatorial microtubule organizing center [IDA spindle pole body [IDA cytosol [IDA gamma-tubulin complex [TAS |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[IMP spindle assembly [IMP G2/M transition of mitotic cell cycle [IMP establishment or maintenance of cell polarity [IMP microtubule nucleation [NAS gamma-tubulin complex localization [IMP nuclear migration along microtubule [IMP cytokinesis [IMP M/G1 transition of mitotic cell cycle [IMP microtubule anchoring at spindle pole body [IMP cytoplasmic microtubule organization [IMP mitotic metaphase/anaphase transition [IC |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRSISSVLA KEESENGSPI PALEEGSVYH VSLKSEGVSP FSDRIQLNYL YDGKTFSDPS 60
61 NLNIHQQEAC LINELLNAFM GMEGVFVHLQ DMKASSEFET IIMPPSFYIL PGFDLGIKDI 120
121 ASEMLEMGSH YLSITAFIES RSHFEYGFVN HALCAALRKF VMDYVVLIMQ CENQSRIDPN 180
181 FSLQTLRLYT LPTSRSLRQV YLILRDLLLS MEKNASSSDD LGLSNIDDLL EQLNEGNDIS 240
241 HVVNATRSKK KVCKGGQVIS FLTESLTKYA GDPVARKILT YLLREASRPY TKMLNEWIHL 300
301 GLVNDPYDEF MIKIHKGITS MQLDEDYTDE YWEKRYVIRE DQVPPQLLDL QNKVLFAGKY 360
361 LNVVLECRKG VNNLASLNAK DDTQNQLLWP STFDDDNFTL NIMNAYVYAN ESLLQLLQSS 420
421 QSLYAHLYSL KHYFFLDQSD FFTTFLDNAQ HELRKPAKYI SITKLQSQLD LALRQPGTIT 480
481 ATDPHKEYVT VEVNQTSLID WLMHIVSISG LEEGTSSQGN EVWNESITKQ ADVGNETRNF 540
541 ESEHNRSTQG TSKVGSDKDI NGFETMQLCY KVPFPLSLIL SRKAIIRYQL LFRYFLLLRH 600
601 VEMQLENSWV QHSKNSAWRL NSSNAKIEQW KRNSWLLRTR MLSFVQKIIY YTTSEVIETH 660
661 WGKFMGELEN ARTVDNLMQE HIDFLDTCLK ECMLTNSRLL KVQSKLLNTC AMFASYTSTF 720
721 TRSLYLLENS EESFDEGRMD KMEEILRRYE DSFSRHLKSL VNACNYFAST ETAALLSLVM 780
781 KLTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
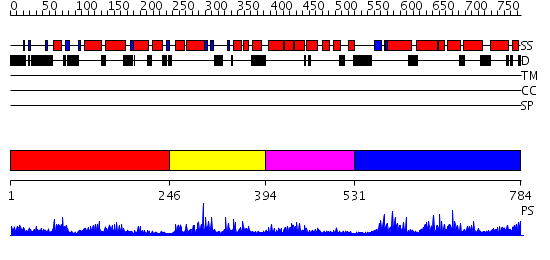
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..245] | 2.14997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [246..393] | 1.18199 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [394..530] | 1.189995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [531..784] | 3.189992 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)