













| General Information: |
|
| Name(s) found: |
FBpp0293026
[FlyBase]
FBpp0293024 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1436 amino acids |
Gene Ontology: |
|
| Cellular Component: |
filamentous actin
[IDA]
|
| Biological Process: |
bristle morphogenesis
[IMP]
epidermal cell differentiation [IMP] antennal morphogenesis [IMP] imaginal disc-derived wing hair organization [IMP] actin filament bundle assembly [IMP] cuticle pattern formation [IMP] |
| Molecular Function: |
actin binding
[ISS][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHSHQLDSYL GGVGSMMNLG GMGQVGQMYH PGSVSGSERP NGGALALHYA AARGCLDCVQ 60
61 LLVAASVDIC ANTQMDNDVT PVYLAAQEGH LEVLKFLVLE AGGSLYVRAR DGMAPIHAAS 120
121 QMGCLDCLKW MVQDQGVDPN LRDGDGATPL HFAASRGHLS VVRWLLNHGA KLSLDKYGKS 180
181 PINDAAENQQ VECLNVLVQH GTSVDYNGKS SSQRHKSQQQ LHQQHQQQQQ QQQQQHLSSS 240
241 CNSNSNSSKQ TSRSNTIKSK SSSTLSSDVE PFYLHPPALA GGSGGSGMGL GMGMGMGMGM 300
301 SMGLGISRKN SDALYSQQSR SSSEKLYNGS SSLGGNPGGN SSSGGGGVGG GGGSGGALLP 360
361 NDGLYVNPMR NGGMYNTPSP NGSISGESFF LHDPQDIIYN RVRDLFVDSD CSSSVKQHGK 420
421 SHAHQLLQSK AGNAMTIQAD VHSSSSGAGS GSDESVSISS SFNSPKQQQQ LRSQSFNRHG 480
481 SSSNISASGN IKNNNYKAHK GKPSNQNGGG DHDYEDIYLV REESRKQHQQ QQQHQLQQQQ 540
541 QLQHQATQQQ QQQQQQQQQL QHKYNVGRSR SRDSGSHSRS ASASSTRSTD IVLQYSNHHL 600
601 NNKRNMNNNS NMNNSSSNIA QSSSSNNNNN SLLNRNKSHS IIGLHSSKYE SSLKDNYSAK 660
661 NVNLKNQLLN GGIKSDTYES VCPPEDVAER TKQTHKNSMI RNNLADASSN NNTSGSINNI 720
721 SNIGNMNGGN QSSRNLKRVS SAPPMQNLAV VNGPPPPPLP PPLRAPVQQQ RNNLSVDQPN 780
781 SMTGNNGIYK KNVFGPNQGP TNANGTGGAA PPPVPAPNPV ATEAVDSDSG LEVVEEPSLR 840
841 PSELVRGNHN RTMSTISANK KAKLLNAGST NGSSIAASSD DSQSRYGGSV HAANGSAANG 900
901 HFYGYSEGNK NQGSNGNGNG NGNGGGGNYH SHPNQPHYAT GQPHQYTPSL YGGGSSAEDH 960
961 YELTQQQQQI YGETHLPSGQ RTGGPNLVNK QLVLPFVPPS FPNKSQDGVT HLIKPSEYLK1020
1021 SISIDKRSCP SSARSTDTED YMQIQIAQQQ SHPHQHQHHP HPHPHPHHPH QLHGMGEQPP1080
1081 LPPPPPPLPH GMLPPQPPMG PPMGVAGGQD ATMSGKSHKP KKPHGSAPNS QDTATRKQHQ1140
1141 PLSAISIQDL NSVQLRRTDT QKVPKPYQMP ARSLSMQCLT SSTETYLKSD LIAELKISKD1200
1201 IPGIKKMKVE QQMASRMDSE HYMEITKQFS GNNYVDQIYL QIPERDQAGN AIPDWKRQMM1260
1261 AKKAAERAKK DFEERMAQEA ESRRLSQIPQ WKRDLLARRE ETENKLKAAI YTPKVEENNR1320
1321 VADTWRLKNR AMSIDNININ LAGMEQHYQQ IQFQQKQQQL QLGNKENHVD PKGADQDQEH1380
1381 GVGIGSGVGN ANSGVIGGQE LIDPSEGSQG RAEEEEDNII PWRAQLRKTN SRLSLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
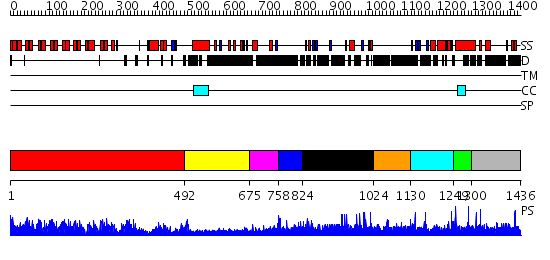
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..491] | 89.69897 | Ankyrin-R | |
| 2 | View Details | [492..674] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [675..757] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [758..823] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [824..1023] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1024..1129] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1130..1248] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1249..1299] | 2.013989 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1300..1436] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)