













| General Information: |
|
| Name(s) found: |
gi|24641890
[NCBI NR]
gi|22832229 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 652 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
purine base metabolic process
[IEA]
purine ribonucleoside monophosphate biosynthetic process [IEA] |
| Molecular Function: |
AMP deaminase activity
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERNDADINF QRVSISGEDT SGVPLEDLER ASTLLIEALR LRSHYMAMSD QSFPSTTARF 60
61 LKTVKLKDRI NNLPVKEVSE SADVHLRHSP MKITNPWNVE FPNDEDFKIK PLNGVFHIYE 120
121 NDDESSEIKY EYPDMSQFVN DMQVMCNMIA DGPLKSFCYR RLCYLSSKYQ MHVLLNELRE 180
181 LAAQKAVPHR DFYNTRKVDT HIHAASCMNQ KHLLRFIKKT LKNNANEVVT VTNGQQMTLA 240
241 QVFQSMNLTT YDLTVDMLDV HADRNTFHRF DKFNSKYNPI GESRLREVFL KTDNYLNGKY 300
301 FAQIIKEVAF DLEESKYQNA ELRLSIYGKS PDEWYKLAKW AIDNDVYSSN IRWLIQIPRL 360
361 FDIFKSNKMM KSFQEILNNI FLPLFEATAR PSKHPELHRF LQYVIGFDSV DDESKPENPL 420
421 FDNDVPRPEE WTYEENPPYA YYIYYMYANM TVLNKFRQSR NMNTFVLRPH CGEAGPVQHL 480
481 VCGFLMAENI SHGLLLRKVP VLQYLYYLTQ IGIAMSPLSN NSLFLNYHRN PLPEYLARGL 540
541 IISLSTDDPL QFHFTKEPLM EEYSIAAQVW KLSSCDMCEL ARNSVMMSGF PHAIKQQWLG 600
601 PIYYEDGIMG NDITRTNVPE IRVAYRYETL LDELSNIFKV NQTCSIADMN AT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
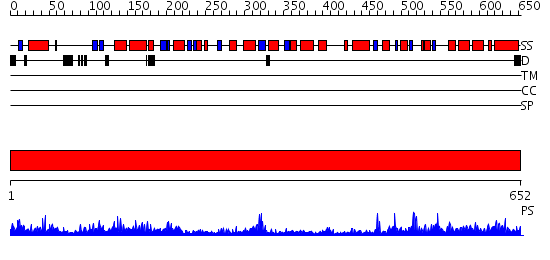
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..652] | 164.0 | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)