













| General Information: |
|
| Name(s) found: |
FBpp0310009
[FlyBase]
CG9940-PA / FBpp0073676 [FlyBase] CG9940-PB / FBpp0073675 [FlyBase] |
| Description(s) found:
Found 40 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 787 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
NAD biosynthetic process
[IEA][ISS]
|
| Molecular Function: |
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
[IEA]
NAD+ synthase (glutamine-hydrolyzing) activity [IEA][ISS] ATP binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRKVTVAVS TLNQWALDFE GNMVRILQSI LEAKDMGASY RTGPELEVCG YSCEDHFREP 60
61 DTFLHSWEVL LEVMMSPMCE NMLVDVGMPV MHRNVAYNCR VAFFNRQILL IRPKMAMCDD 120
121 GNYRESRWFT AWTKALQTEE YVLPRMIAQH TGQQTVPFGD AVIATRDTCL GYEICEELWN 180
181 VRSKHIEMSL AGVELIVNSS GSYMELRKAH ITSDLIRNAS FKAGGAYLFS NLRGCDGQRV 240
241 YFNGCSAIAL NGEILARSQQ FALQDVEVTL ATIDLEEIRA YRVSLRSRCT AAASAAEYPR 300
301 IHCDFEMSTH SDIFKTSTPP LNWPMHTPEE EIALGPACWL WDYLRRSGQG GFFLPLSGGV 360
361 DSSSSATIVH SMCRQIVQAV QQGDAQVLHD IRQLLADSDY TPDNAAGLCN RLLVTCYMGS 420
421 VNSSKETRRR AAQLANQLGS YHIEISIDSA VNALLSIFNA VTGLTPRFRT QGGCARQNLA 480
481 LQNMQSRIRM VLAYIFAQLT LWVRNRPGGL LVLGSANVDE SLRGYLTKYD CSSADINPIG 540
541 GISKMDLRRF LTYAKDKFNL PVLESIIDAP PTAELEPLQE NGELQQTDEA DMGMTYAELS 600
601 QFGRLRKQSF CGPYSMFCHL VATWKSDLSP KEVAEKVKHF FLCYAINRHK MTVLTPSVHA 660
661 ESYSPDDNRF DHRPFLYRPN WSWQFKAIDD EAEKLQPIYT PSSAQLRPSS EDLLISTQRS 720
721 SHLDDSKHSS PLSSASASAS IDVGISTAAV PLPGAAAPGG LSKKPSGYSK VHVNVLGKIK 780
781 DRTGIPV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
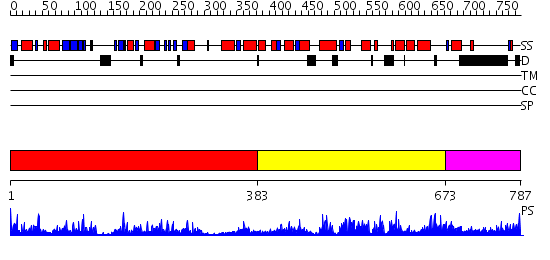
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..382] | 56.0 | NIT-FHIT fusion protein, C-terminal domain; NIT-FHIT fusion protein, N-terminal domain | |
| 2 | View Details | [383..672] | 60.221849 | NH3-dependent NAD+-synthetase | |
| 3 | View Details | [673..787] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)