













| General Information: |
|
| Name(s) found: |
CG42237-PA /
FBpp0073553
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 363 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extracellular region
[IEA]
|
| Biological Process: |
anesthesia-resistant memory
[TAS]
lipid catabolic process [IEA] regulation of heart contraction [IMP] olfactory learning [TAS] phospholipid metabolic process [IEA] |
| Molecular Function: |
phospholipase A2 activity
[IEA]
calcium ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLIQNRWQE IGLQLIFVFN FGWIRRRCFW VAAQSSRRCL SGSSSRSKKP RHTKMPYNLW 60
61 LRRRLQLLAS LLFCCLLGYL TSEAVAKPTL SFSLPQGLQG FPSFQSFTQQ IIEQRSARQM 120
121 KTYSGKRVSN DSLIMIYYHD LTIAVTELGP QKLLLGCELI EIYNDKEGKM LLEGLSHYNR 180
181 PLEIKFDEML KLMDQCEHVD KLSYASRHKS KLEGGERSNG GSASATTGGA NDGVALKLAT 240
241 NIFPRSPFSL LSGIIPGTKW CGTGDIAETY SDLGSEMAMD RCCRQHDLCP IKIRAYQNKY 300
301 ELMNDSLYTK SHCICDDMLF SCLKMTNTSA SQLMGSIYFN LVQVPCLDGR SNHYKFRAAK 360
361 EGF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
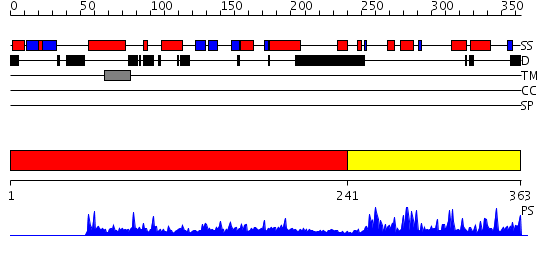
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..240] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [241..363] | 39.221849 | CRYSTAL STRUCTURE OF BEE-VENOM PHOSPHOLIPASE A2 IN A COMPLEX WITH A TRANSITION-STATE ANALOGUE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|