













| General Information: |
|
| Name(s) found: |
FBpp0310024
[FlyBase]
mxc-PA / FBpp0071267 [FlyBase] |
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1837 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
hemocyte proliferation
[IMP]
hemopoiesis [TAS] hemocyte differentiation [IMP] |
| Molecular Function: |
DNA binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESIVLHSDV ARLVLGYLVN QNLKRAAHTL CRTSPHLRHE FLALKQGLQT HNFLHGGLEE 60
61 IICEHVKITS LVAGAVQKLP LDMRMQLQQM KLSERVSELI AAGERSSCTN ESSTPGEPTI 120
121 SQSHRKRRRL RTHSPVNSIS SPSFSKRPRL LPPHFYCSVN RDKVRQSFLG SQDDCEAELD 180
181 EEDGESTTAT EDLEEDDLLP ANGPGPRNNS TPRQGHTESN PLLLTPQSMP ELACAIIKNQ 240
241 DFQETLVKNI NVALQTVTVN NPSVSCDAML DGLVKNILEA TEKDPSFDRI IQEVVIGDEP 300
301 LANEEDPGVS PASDAAGAAI VPAADGDVLP TEQAVDPVPP QTPLIIRTAV AATAGTNADP 360
361 NFSISKLIVL NSNESVQKMV AGLDTSNISF TSEGLNINFA DPASMLSEVE ATAAGQVCVD 420
421 ATTGQLTFPM YLSNGGLLSH LPFLVNNEWV AQQLGPDAFA NIDDSHIEIS LPEPITLSAN 480
481 QLPPNSIIIS SAQKQPPPPS VEPPVQLIKD AAKVDNKAGQ GTAAKEQPID QLEEEQRKFP 540
541 PASLDSSRQV MERVTPSTSG IINVKAFRSL STPRKRTSHV RTLTFSPKVP PGGVLHSNTP 600
601 LSTRRPGLLA KAQEKPIINH VEIIQAACDL SSNEGLGGVP PLFAMEECSN QTVIRAHPPA 660
661 AAPSAAPVPP LVATPKRKQK RQAAVKACKR IISQAESETK TDKDQKKIMP KNNSACSASS 720
721 TAHDDSAEAS KENLQEQQKI EGKPDVGSAA RGDAVVDNDM AAWQRQLHGS NSDLENRLRE 780
781 INSKREEVKA SGRSRRPKKK DTPTSASSRA RKAPMVKAKA EPMNLAKKPQ RSVGDSTEKI 840
841 NIKIITPQKP KLLKKKKVFE AKEEIIDETI EETIDETQAE IEEKDQKKKS EQGCIYAKDQ 900
901 EQAKALAAEL SRPNMAMLLD TPFKASPIGA GASGAVEDDT TSKEAGVGVA AIIPPTPGMA 960
961 IPPLDTPYGK LPSSSFLFGS DTKSIMDTPQ LSAITPGFRL TPFGQMPGTP IGSAAKTEYS1020
1021 SGSSYYRPDE AEHTDANAQC AIQQWGDPQD KKQPKVRSTP KKKKVEAEDE LSQQKNQELI1080
1081 EMECSKKSLH PEDKHAEDNK KTPKRPEKPM DEHLKETEND GEKNETVLEK TPDAVKQPVV1140
1141 LSLEPTVLRR VRSFGSEAVD SAESATAGSF PSDQQQLPHY KLLPGLPEAI IEGSSNSSSS1200
1201 ATSSSSSSSS SSSSSSSSSS SSSSSSSSHS GKGESRGEEA NGENADEKCS KKKEMLLNID1260
1261 NLSNISSTED EEWLKAAGSE DAVLPMLAID SQPGQLVSQD GEVRYPIRNW LTPSKEQAQE1320
1321 TDTSGEQQTK PTPPKPVAAP VPPPPLPLTP PAAPPPPPPP PPEADDPSSV ALPTASTSRQ1380
1381 EHQRQQLNEK RQRVVAKFKE QPQPIKGQGR KTANKKLTAM RENAKLANPS SQKSPKNRKH1440
1441 IEPLKTKAAK KQIEPNPNVK STEATGPPVG AATDAPADPP AGKLDPALLM ALNLSAKKQE1500
1501 SLPIVIKPKV NRAAVEIAAQ PAPGKRGRPK KVQNGEPANL CMRRSSRLVE NKTPAPEDLS1560
1561 KSTEGAKTKT QIKANTKKPA KKRVEARVPS STTDAPALPI VAEAESTQKS PKQKPQIQQT1620
1621 IQEPKQDPPP SKCDSEPDDS DFEQDMCLSS SQERFTYNFT YADNGPKSSN PMIRQPSSYF1680
1681 KSYQMRLMTD DEQLHTLRIA SAQVLCLGQP VASAIRNIPK KRIRRLTGNS GGTAATGGSA1740
1741 SVSGGSGTGA EVAAAGEQPS ATSTPLVEKP PSRSGPQDEE DLEVATINSA PIAANSSSGI1800
1801 GGEETPENSA QQKELQSENH IVEFEDIESI LSHLHGT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
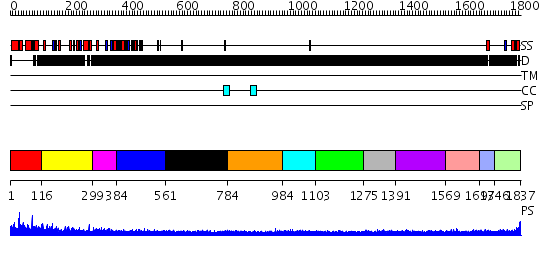
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [116..298] | 2.173996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [299..383] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [384..560] | 1.189997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [561..783] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [784..983] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [984..1102] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1103..1274] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1275..1390] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1391..1568] | 2.146994 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1569..1692] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1693..1745] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1746..1837] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)