













| General Information: |
|
| Name(s) found: |
dalao-PA /
FBpp0071235
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 749 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
brahma complex [IDA][IPI][NAS] chromatin remodeling complex [ISS] |
| Biological Process: |
chromatin remodeling
[IGI][IPI]
positive regulation of transcription, DNA-dependent [IDA] |
| Molecular Function: |
DNA binding
[ISS]
protein binding [IPI] transcription coactivator activity [IC] general RNA polymerase II transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALPSNYKQI AVGGQGSATP LQGGGGGSGG GSRSRSSGGG GGGDRNKDQT PIFTHSNYGN 60
61 PAFTPQKVTK SSSSKNQNES RLPKPPKPPE KPILPYMRYS KRVWDSVKAK HPELKLWELG 120
121 KKIGAMWKLL PEDEKTEFID EYEAEKLEYE KSLKAYHQTP AYQAYMSAKS KVKTDVDMHE 180
181 TPSRGGGSKS QHERRIDIQP AEDEDDQDEG YTTKHLAYAR YLHNHRLINE IFSEAVVPDV 240
241 RSVVTTTRMQ VLKRQVSSLT MHQTKLEAEL QQMEEKFEAK KQRMVESSEA FQEELKRHCK 300
301 PAVDEETFQK MVLRMYEDIK RDRQRLDEPN ANANSAANPA AAAATAAAAP VTRSEEAVKP 360
361 PTQPGQPAAT PAGQEPASAV PAPAAPPKET PPAVKPATLN PTPSSTPTPA PAVHVHETAS 420
421 KTDPEPMDIE PPPKPSVPPP PIKPEKLEMA AALPPQSTLV EPPKTEPAKV VAQPGKVPTP 480
481 VPTPTPPPEV APAAGAATAA TTTTTTTPTP PPVPQQVPLP QQQGGPAPPP GMPQMHPHPG 540
541 HPPPGHSLMP PHMGPHQPPP GMPGLPPPPP HTGYANYGGP PHGPPPGPPG GPARPYYQPQ 600
601 YGGHPTPQPY YAPFSPYQQS YGPPPGSHYM SPRPPPPQHN GNPGHPYAPE HGSNPPPPQQ 660
661 QQQQQPPPGH LHEPSGGGPG APGGGAGAAA AAAPGAGVYP TPGAGAGPGA PPGPAGGAPL 720
721 GEAAVAGGVA PPAATAPGKH PEAEKHEAD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
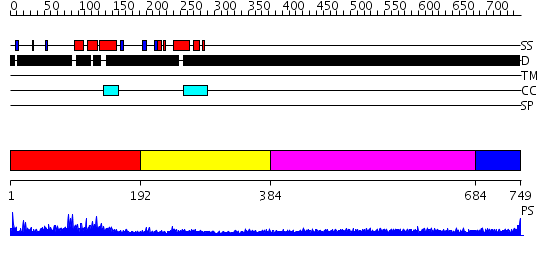
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 18.0 | No description for 2gzkA was found. | |
| 2 | View Details | [192..383] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [384..683] | 2.247994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [684..749] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)