













| General Information: |
|
| Name(s) found: |
CG32758-PA /
FBpp0070808
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 531 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[IEA]
|
| Molecular Function: |
protein binding
[IEA]
phosphoinositide binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIVGENHPL PTPTPASAVV TASSGGGGGG AVVMGVGVGV TTANGPRVVT IYKTETGFGF 60
61 NVRGQVSEGG QLRSINGELY APLQHVSAVL ENGAAEKAGI KKGDRILEVN GVSVEGATHK 120
121 QVVDLIKSGG DCLTLTVISV TQQEADRLEP QEDQSGYSYI DYSDKRSLPI SIPDYGIVNR 180
181 NGERYIVFNI HMAGRQLCSR RYREFANLHS LLRKEFSGFN FPKLPGKWPF QLSEQQLDTR 240
241 RRGLEQYLEK VCAVRVIAES DAVQDFLTDT EDDISASPVD IKVMLPDHEV STVSVKKSSN 300
301 AQVVWEILVQ RANLTAYTQQ YFYLFEIVEY NFERKLQPHE IPHQLYVQNY STASSTCLCV 360
361 RRWLFSVAKE LTLPDGEQAG RFIFYQAVDE VNRGNIRADG RLYELKALQD AKKAGDYLAL 420
421 ARTLPGYGDV VFPHCSCDSR KEGHVVPAVG MKSFRLHACR EDGSLEAQMV ELTWDSITRS 480
481 ESDEESMSFC FQYNRPDKPA RWVKVYTPYH AFLADCFDRI MEERKWEDSG D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
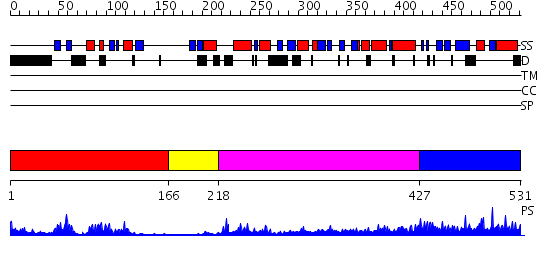
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 17.522879 | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | |
| 2 | View Details | [166..217] | 6.0 | Psd-95; Guanylate kinase-like domain of Psd-95 | |
| 3 | View Details | [218..426] | 17.69897 | No description for 2dybA was found. | |
| 4 | View Details | [427..531] | 1.043986 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)