













| General Information: |
|
| Name(s) found: |
CG3626-PA /
FBpp0070620
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 939 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial pyruvate dehydrogenase (lipoamide) phosphatase complex
[ISS]
|
| Biological Process: |
glycine catabolic process
[IEA]
|
| Molecular Function: |
oxidoreductase activity
[IEA]
[pyruvate dehydrogenase (lipoamide)] phosphatase regulator activity [ISS] aminomethyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWKLLQRSGN AVSVVPRRRW CSPGLSNAVR LSSSQMDPAD ERVLRKRKFQ PQQAADLSEE 60
61 LAGQLPVKSR VVICGGGITG ASVAYHLGLR GWGGETLLVE QDRVGGELPW TACGLAGRFE 120
121 PSYTELKLAE YSIDLIKRLA ENGLPTGWRP VGSLNLARSW DRMTAFNRMK SQALAWGMHC 180
181 EILSPEQCAQ HCELLSLDGI EGGLWIPEDG VCDPQLVCQA YMIEAQRLGV RIVEHCAIKK 240
241 IHSEHGKVRS VETTAGDVEC EYFVNCTGFW AREVGTLSKP VVKVPLKAVE HHYLHTKPIE 300
301 GLSPDTPFVR DFDGRIFFRE CEGHILAGGF EREAKMVYED GVIPLSQTAR QHPPDWDHFH 360
361 ELLDALLLRV PSFRDATLDR LTNSLQVFSP DCKWILGEAP EIQNYYVAAG NKTMGVSASG 420
421 GIGRVLTDLI TKGSTYLDLH ILDISRFLGL HNNRKFLRDR CKEAPGKHFE INYPFEEFQT 480
481 GRNLRMSPIY PQLKEAGAVF GQSMGYERPN YFDQQDKHDE FGLPRFRIAQ TRTFGKPPWF 540
541 DHVASEYRAC RERIGIADYS SFTKYDFWSK GNEVVDLLQY LCSNDVDVAV GSIIHTGMQN 600
601 PNGGYENDCS LARLSERHYM MIAPTIQQTR SMCWIRKHMP NHLRAKVNVA DVTSMYTAIC 660
661 ILGPYSRILL SELTDTDLTP KSFPFFTYKE LDVGLADGIR VLNITHTGEL GYVLYIPNEY 720
721 ALHVYSRLYQ AGQKFNIQHA GYYATRALRI EKFYAFWGQD LDTFTTPLEC GRSWRVKFNK 780
781 PIDFIGRNAL LKQREEGVKR MYVQLLLNDH DHEVDMWCWG GEPIYRDGVY VGMTTTTGYG 840
841 YTFEKQVCLG FVRNFDDEGR ELPVTNEYVL SGHYEVEVAG VRFEAKVNLH SPNLPTKFPD 900
901 REREAYHATR DKPDQADLLS YGGVTTVKGT GIPTDGHGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
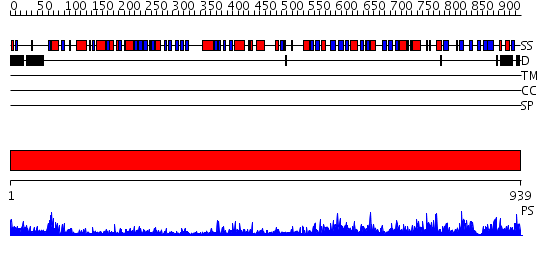
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..939] | 150.0 | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with acetate |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)