













| General Information: |
|
| Name(s) found: |
GlcAT-I-PA /
FBpp0070612
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 306 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
plasma membrane [ISS] |
| Biological Process: |
proteoglycan biosynthetic process
[NAS]
|
| Molecular Function: |
glucuronosyltransferase activity
[ISS]
galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity [IDA][ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEVRIRPRQ VLILIIVFLV VLMMVHRNGK RTCQGPEYLQ AMFVQGDTLP TIYAVTPTYP 60
61 RPAQKAELTR LSHLFMLLPH LHWIIVEDTN ATTPLVRNLL DRAGLEKRST LLNIKTPSEF 120
121 KLKGKDPNWI KPRGVEQRNL ALAWLRNHVD VDRHSIVFFM DDDNSYSTEL FAEMSKIERG 180
181 RVGVWPVGLV GGLMVERPLL TEDGTKVTGF NAAWRPERPF PIDMAAFAIS MDLFIRNPQA 240
241 TFSYEVQRGY QESEILRHLT TRDQLQPLAN RCTDVLVWHT RTEKTKLAAE EALLKKGQRS 300
301 DGGMEV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
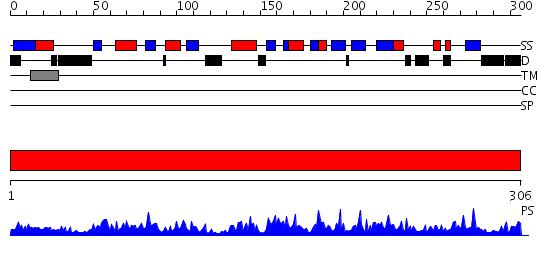
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..306] | 100.0 | 1,3-Glucuronyltransferase I (glcAT-I) |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 |
| Source: Reynolds et al. 2008. Manuscript submitted |

|