













| General Information: |
|
| Name(s) found: |
FBpp0310121
[FlyBase]
Mur2B-PA / FBpp0070274 [FlyBase] |
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1795 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extracellular region
[IEA]
chorion [IDA] |
| Biological Process: |
chitin metabolic process
[IEA]
|
| Molecular Function: |
chitin binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTHCLWSTLL GLLSVLSTAT SRSVRQLPSA YPPVLPTGYY GQPPYAYPPY GYGYQAPPPV 60
61 YQPYYDNVFG VFKSYPGGGF PVRFDYNNRC SRNYIGIKPH PDQQQYYYVC KPDCVIFSKC 120
121 RGLESFNASS GRCVQHVPQH RPDHRPPQCQ KEGRFPHPHD CKVYYRCDKN RTQPWLFACP 180
181 AGTIFSPVER KCLPGDQCPS TEISDSGSYI PQNCELKFPE CAEEGTFRSP TDCALYYTCR 240
241 LQESGTYLQT RFKCPGSNSF DLERKLCRPR SEVDCFDFVP GPVQVPYAPQ PYYPPYPAAP 300
301 PLYEEDDYDT GAREQQPALK SEKLQVAAEG FEKPSLNVVV LQTTTLEPST AYHKYPAYPS 360
361 YPSYEYSSHH RGKERAAENL ELEKEGVPRK LKLSENIVIQ PETPATAATT REPLNDINKY 420
421 QYKRYTYGTD KNDVTEAPEI KSPLKGLHLS ENIVILPETT TTTTTTTKPV VLTCPTISPP 480
481 DTTPKPSTTT AVTKSTPKIS STEQHSTTTA KTTTTKRPTT VTEKTSSATE KPRTTVVTTT 540
541 TQKRSTTTHN TSPDTKTTIR STTLSPKTTT TPSTTTPSTT TPSTTTPSTT TPSTTTPSTT 600
601 TPSTTTTVKV STHRPRTTSQ KTTTASTTTK KTTTSPKTTK TTDIPTSTTS KLSTTTQKTT 660
661 TTTHKFTAAT TSTEKPKTTT EKTSTVSTTT KKSTESSPKP TSSTGKPTTT PKPSTRTTPT 720
721 TTKVTTTTQI TTTTPLRSST ETTSTQPPTT TTPQPTTTTT LTVTPKTSTT TTTTEKPITS 780
781 SPKPTTTTQK TTSTAPNTTK VAITTQKETT PTQSTSTTIF TRKTTTNNPE PTSTEKPITS 840
841 TTPKPSTTTP KTSTVASSTE KTTISSPKPT TEKSTENPTT NSVKTSALTS STQRATSTTS 900
901 EPTKTTQNIT TTTPKPTTLK TSTQEATTST QKVSTVTITT KKATESSPLT TLSTEEPNTT 960
961 PKPLRTTTPT TTSVTATTRI TTTTISESST ETTSTQKPKS TTPTSTTRTT PKVTTVIVST1020
1021 QNPTTTTSKT STVTITTPNP SPSTQRPTTT TRQPTSITAS TTSIGTTRIP TTTNPQNSTS1080
1081 STDLTTVTRP PCPDPDSTSD KNTNTACTQE LQQVNLLELQ SPQKQEQFTH TRTHTALTGS1140
1141 RNTLGGQEVP DYMDDAPSSA EAESGQATTA KAPTMSTLAA AHLLQKLFHI ISTTPPSREH1200
1201 APTQRPSSQP SSSQRSRGVT IAQMARHNLA TSKPFIAHSL RLSIQQLAST QKRSIPPKTL1260
1261 VTHNTTKEPE DSEYYDSETS EQYTDEDNEV LDKTQPRAMS STTVAAVLPA VPSTTTEREP1320
1321 QKTSSSPSPT KATSSTTTQP IETTTGDLEY DSSGSSDYVN DANDISSGVV NSLEARKNFL1380
1381 LSLLKQRLTQ IERTEAKKPA TSTSTTTDAP KTSSSSTSPA STTSESTSPV TSSTARSSLT1440
1441 ASKHLGPEAL SRQKSLTPQS AEYYDEDDDY MEDEPVGSSD AEKKEHGTVL ISEKQAAATA1500
1501 KRHIAPPSAQ PLRQAMLNIL ATRSEENKVD LKTTSDGLQF RRFESSQIPT EGPPNRASIV1560
1561 VAEALSRRLA DNVGEHLPGP TVPHTSDAKA ISSVSQTTIR SRESQTKSSS QTPPSISTET1620
1621 PATQAIQNHR TDEATQSTLE IVTQTTPKSA PPATAVPVAV RQEVLSVNSR PWLLAARNQT1680
1681 VHLTPISSIA ARAPSNPVSH ANRSINPLVS SPEDHSTEKV PQLIVKVYEP IDLKIVFCPK1740
1741 SCDQDHDHKF DPADNIKTSN CTGGCDEEEE RIHKHVDIQW KPLELTDIAG FRNAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
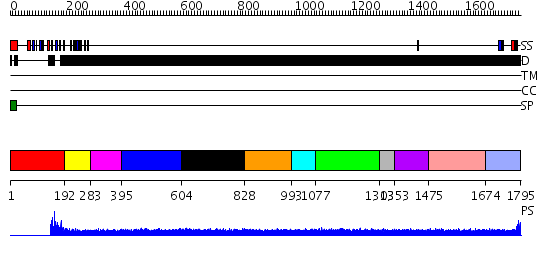
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [192..282] | 9.045757 | No description for PF01607.15 was found. No confident structure predictions are available. | |
| 3 | View Details | [283..394] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [395..603] | 1.118982 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [604..827] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [828..992] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [993..1076] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1077..1302] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1303..1352] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1353..1474] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1475..1673] | 1.102983 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1674..1795] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|