













| General Information: |
|
| Name(s) found: |
gi|74867920
[NCBI NR]
gi|7302107 [NCBI NR] gi|24651778 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1844 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
signal transduction
[IEA]
|
| Molecular Function: |
protein binding
[IEA]
GTPase activator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVQPAEMAE NGRSVPDVTA SPGRAPPGPL PANQMPAMGN QQHHGNQQHH GNQQQHHGNQ 60
61 HSNHRGQSGS LSNAAGVKDP VMLQGDFRKV SGISSEIFRQ IEAVENDHDP NTAAALEAVE 120
121 RRGEMIVRVL EPRCMGSKQA VDAAHKLMNK ADARHTVQLV EIVKRPGQTL GLYIREGNGA 180
181 DRTDGVFISR IALESAVYNS GCLRVGDEIL AVNLVDVTHM SLDDVVIIMS IPRRLVLAIR 240
241 QRRGNRGTGS PGPPTLSRPE QKPPPVVVIK RDLRDEDLDE TDRMPRPRSS RDGREMTESR 300
301 SRLGLGLNNY SPQSEQLDMY YNTRGGGGGA MGEPPNWGYK PPPPPSSVIT EQPTKAHAFA 360
361 PSHAYYQNAG TLESLAEKVH AFYPGQPGGP PVGPSRRMST GTGNVGLAQQ HARFPRSGSD 420
421 QHLPRVEYSD YSNSLGRHSL LRSSLKPGTT GGAPMQVGVG GTLGRYGRYD QQRAGVSKYG 480
481 PPSGGAQSLT RRSRPNLDYS SDTEATIGPR PSYYYYNRPA IGSMSRGSGG AGGGVGAAST 540
541 AALLAGAADL NKFNSLPRER PGTRLQGIRS RMGDRLVDEN DGNTSAPEFD VRRGRDLRQR 600
601 ITASPSIFTA DEYRAWLRRA PSSSAIAEQM RMTRDMFAQP RAQRFSCSAE NIHDALRNTE 660
661 SIYSSRNHIL GTGTLDRNMG LTRPISALPV RSMSSQHIGG AGSIRSPSIR RMRQLLELSA 720
721 GPASPSGSIL STGGHQSPAP TPSATLPRPH RQIDINPAEF AKYKLDKPIV DIGGISGMLW 780
781 IHLLAGRGLR TAPEGAAGTA TQGQTRDLYC VIECDRVHKA RTVVRSGDLQ FDWDESFELD 840
841 LVGNKQLDVL VYSWDPQHRH KLCYRGAISL SSILRQSPLH QLALKVEPRG TIYIRMRHTD 900
901 PLALYKRRGL PSLRAGYPTL FGADLETVVN RESKNAPGSA PVPIVLRRCV EEVERRGLDI 960
961 IGLYRLCGSA TKKRLLREAF ERNSRAVELT PEHVPDINVI TGVLKDYLRE LPEPLFTRCL1020
1021 FQMTVDALAV CLPDDPEGNA KLMLSILDCL PRANRATLVF LLDHLSLVVS NSERNKMSAQ1080
1081 ALATVMGPPL MLHSASAQPG ADIDHAQPIA VLKYLLQIWP QPQAQHQQMA QHMGGAAGAM1140
1141 MGGLVTAGSM SNMAGVASGN TGRRGESTGQ RGSKVSALPA DRQQLLLQQQ AQLMAAGNLL1200
1201 RSSTSVTNIL SQGHPQLSAT ANNHLYQSVV GQLAQSHRAL QQAVQQPYQL GGSVGSAIPD1260
1261 PSPLPLPGTP SPGSSSASTG SGSGSGKSTD TIKRGASPVS VKQVKIVDQP SSPYSIVMKK1320
1321 PPLQKDAPVE ITTPTTQADT ESTLGCKESN GTASRRGNVD FYDTHKTQAK SVVNEESSYS1380
1381 SKYTGSETKK IIPGNSSYTP SKANASGLSG GEDYKAMRNK SSATSSSSSS QATVLSAGST1440
1441 ATSAPTTSSD DSDDLVSYKS SASTNALLAQ SQAMTTSQLM SKYLKREPRV QFTPIKSPES1500
1501 PSPPGSGDGL PKGTYQLVTP ISGSSSKPGA TTGAISKYTT GSVESSINAN SQKLSSPSRL1560
1561 CNSKDSNSRT GTASSTTPAT SMVSTGRRLF DSLASSSSSE TETKTYIGGT TAASGAITTT1620
1621 IYTNDTKNSG SSSSKSGIGG GSGTGLGAVS GASSETRSFG STLFGSSGLG NGNGSSHNHS1680
1681 SASPSPFTTT NGNGNHNTMH LYGTLPKNGT STGAALFGGS ANSSSYHSSA SGSGAGTASS1740
1741 SGVSSMTGST NSYDFYTSTS STVSSSRPFA NGGNNYHTLG TYRAQYAATN PFLDAFDEKP1800
1801 GSNGGNAHGE EKLGADKGHH RAAVMAFQSS GDSKNGSDEY DDIK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
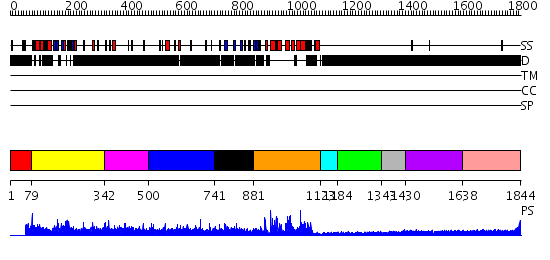
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [79..341] | 13.30103 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 3 | View Details | [342..499] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [500..740] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [741..880] | 13.522879 | No description for 2j59M was found. | |
| 6 | View Details | [881..1122] | 56.522879 | Crystal Structure of the Human Beta2-Chimaerin | |
| 7 | View Details | [1123..1183] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1184..1342] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1343..1429] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1430..1637] | 1.206992 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1638..1844] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)