













| General Information: |
|
| Name(s) found: |
CG1815-PB /
FBpp0085184
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1121 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
protein kinase C binding
[ISS]
zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESTDSSDSS GDSLKSIPRP DFEALSAIGS PQPQLQSSSL DGHVEMEPVI MPSKRQKYIV 60
61 SAPPGSANCS SNSSSSSPSN INVGSSLTMK LTKVQPQQHQ AKSSTPNNKG KVAKEMVALQ 120
121 RSHIESEVLS NFVTGVSKLK SRKSRYVPGN LATQAVGGEL SRSLNTSSEN ENKSLKVALK 180
181 RSSSIPAPIL DSDDDWQDED IAWGVTTCRP RGRSMAFEDG ERLANLNAED RNIVPAYSMA 240
241 TRCRSRSKTL SLSNSWSNDD KGPRRSNLRS ENEEFAKKHN AFLDRIIHDD RESELALNTS 300
301 GEGESSLFSD ASDTKTTTAK TPEEEAAERL ALIYEEPPTP GWDPFCWKCR GCGKLMPCSK 360
361 CLRSFHSYCV RPATTKFDSS WKCPECQVIE AAPKRLRRNG VSVDLLSQLL SFALDRMKHV 420
421 RGAHKLRSPL EVFPLTYKKF FVNPVSFESL AQCIRNGAYQ STDEFLSEVK WIQHNALILD 480
481 AGDAKVEQAS KAVVKVCRQE ANEIDTCPEC YLNANSSDEW FVKVCRHPHL LLWAKLKGFP 540
541 YWPAKAMGSS NSTLVNVRFF GKHDRAFVPV KDCFLYSAQN PNTQTSRRSA RDLAECIREV 600
601 EIHIDHIKRK IGAFNYAPYR TPYDPLEEQQ QLEQMMPGVY AAIDRELEPA NKTPLQFLIR 660
661 KTADDKLSIV KKTKATESGN ESDQSPSPTK KLSEVDVVSV TGSGCSDHSN VKSNNYEVIS 720
721 RSGESLTDSR CKVLLKRKSL AAKIVAESVE TSEAVAPKRK HSLSDASFTS ESSEHKRKSK 780
781 HARKQHDNQD NQIEEAEKTG QEPPKSPTIS KQNENLRDEE NVIENAANDT SSASPVSASV 840
841 VSVVELVRRR QGVTITKIPR EQQQTAEDTA AVPIPLPTAP PPKQNIPKGN EAANPKQSDV 900
901 ERQQEQLIKK VIPFIEIKTE VMSEPDDEGI EEASPANQPQ TDQVPLQQET ITAQPESQMP 960
961 AAAPAPQPKP VDNAPFEEVR IKEEILSEDE METEQSIVSR RLKSVPPMPL PMPPPPPLPP1020
1021 REESPATADS VRFVGDTTIQ RVYPRHPLQL TRPFCQPHLH LRHHQSQLAR LLLASHRLRH1080
1081 RPRESLRHTF NFEEKTIVSQ PPHRRLLHQP QTRLLCSDPT W |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
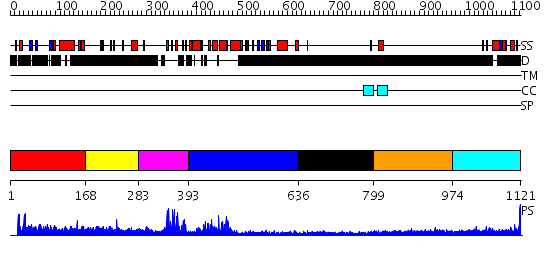
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [168..282] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [283..392] | 8.522879 | No description for 2ysmA was found. | |
| 4 | View Details | [393..635] | 21.30103 | TAFII250 double bromodomain module | |
| 5 | View Details | [636..798] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [799..973] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [974..1121] | 3.254993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)