













| General Information: |
|
| Name(s) found: |
FBpp0305956
[FlyBase]
yata-PA / FBpp0084843 [FlyBase] |
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 873 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWSFFSRDSS KDFPYDIGEP VGGFDQYSIW TLHKAKRKTT LEEVSVFVYD IRSGSDTKCE 60
61 LAKAALKRLK TLRHPSILQY LDSLETDKML YVATEAVDPL GTYFSKLGSD NVQKGLYLAW 120
121 GIFQITRALS FLNNDGNLRH NNVSAWSVFV NASGEWKLGS LEYVSAADGN PMPPAKIPVT 180
181 LEVYDSPEKN DPSKLKAATK CSVDMWGLGC LVWEAFNGVL KQRSNLKDIE HIPKSLQSLY 240
241 CELVGASPSN RPNPADIITR CRKPGGFFKN DLVDTLLFLE EIQIKDKAEK NRFFSGLTTH 300
301 LDNFPDNVCR HKILPQLITA YEYGDAGSAV LAPMFKLGKL LDEVEYQKRI VPCVVKLFAS 360
361 TDRVTRSRLL QQLDLFIAHL QPQVVNDQIF PQVAHGFLDT NATIREQTVK SIIHLAPKLN 420
421 YNNLNVEVLR HFARLQARDD QGGIRTNTTV CLGKIAPHLH PQVRQRVLVS AFIRAMRDPF 480
481 PPARVAGVLA LAATQQYFLL SEVANRVLPS LCSLTVDPEK TVRDPAFKTI RGFLGKLEKV 540
541 SEDPSLRETM EADVHTATPS IGNAAATWAG WAVTAVTAKF YRSQSDSSRP RPPLTGRNLS 600
601 KPASLEQPSS SSLSTTTSSV TSMTSLEHES NDTSASASDY GNNDWDNENW GEMDTSQDPS 660
661 SPLAASSNNG ALMAANALSE VRDGWDNEEW GSLEEDPCEE EEQAEQEQQQ QQLARQSISS 720
721 TSSSQPNQRP LLPHQQQYPH QQQLQQHELN DLIEPLAKLN SHVTSPSKQS MLRPKELPNL 780
781 SSSNTSPTSA TANCNNISPP SSHLNNSTHN ANWNSDSWAD GEFEPLDESG FGNAKLDEAR 840
841 RKREEKKLQR QRELEARRAQ RASGPMKLGA KKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
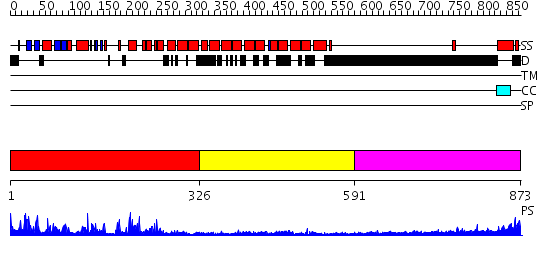
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | 77.09691 | No description for 2qnjA was found. | |
| 2 | View Details | [326..590] | 3.15 | No description for 2ie3A was found. | |
| 3 | View Details | [591..873] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)