













| General Information: |
|
| Name(s) found: |
Brd8-PA /
FBpp0084763
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 872 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
negative regulation of gene expression
[IMP]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAGVQERLQ LSRMPLDTWS KREQLILASA VSCSGDQNWI TVSRTLKTIC GNGSSNRPAD 60
61 WFSQKNCAMQ YGNLLESVEA TKRKKRSSES SAGVSSPATV ETPTELLLRR LTEERQAEIK 120
121 AQMRQDQETY RRIQREIESL QSDAVTEQEL QDMWLEIEKE QEARRIEEMK LENRMREREQ 180
181 RKKDLASNWR NSSLAVKRSN QAADTTSVDM DVEDINTSGN GKQQTGPSPL LTSLLKSPTG 240
241 NAPATPTPGG AAPAAGGTGS VARATAPTIT SLLTSGPSSA VANQPVITMK SPTDAAFMSR 300
301 PITGPPASEA STTPTLERSP SQAAPTLSML LEKNMAAAKA NESGSLVKPE GSGNQEQGVD 360
361 EATSSAAGTA DSDEADPNEA QLLEVFKNID QIIDDIDIDM ASVIDDEILK NVDDVVASPS 420
421 NDKAEVIDFE DKLDALKREQ SKSSPASDKP KSVELVIGSS DDSNDNIPLA AVASQESKDR 480
481 GQSGGESTRG TEDAESEPLT SEVAVEEIGE TITIISTSSE DTAGSPPTKM EEDGATDSLP 540
541 KKEDAAEDKS EQESSSQGQN KAGGSTDEPE LIDHQATREA QSKSGKDTAG KPVTMQEKTT 600
601 TQSAPVPVDL HDTDEESSSL TDSSKHDDHP RSAKAKPPAL KLALDDSKSE LELSGTPQPP 660
661 SAPARTPLLR KLPHRDRSES PMVDDDATTA SDHSTARSTR RRCSSTPVID SIPNSPASSE 720
721 HTDDRRETRA ASKKLFLSIY AMLLDSKHAA PFKRPFHDEH AQRHADLCLR PMDFPTIKRN 780
781 IDSGFIRSLS ELHRDVLLMA HNVLVAYKPH TAQHKTARLF VQDCQAIKEF SQLPDVQAGI 840
841 TATPVSNTGS LRADKSSGSK ARSGSRKSQR HH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
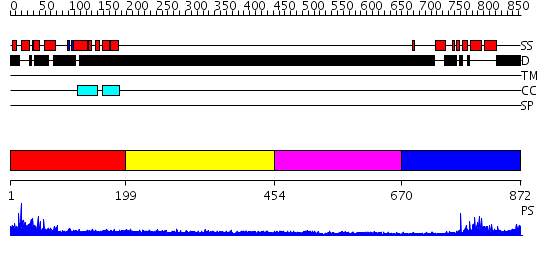
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] | 4.291997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [199..453] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [454..669] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [670..872] | 25.69897 | No description for 2f6jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)