













| General Information: |
|
| Name(s) found: |
CG5508-PB /
FBpp0084620
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 757 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial outer membrane
[ISS]
|
| Biological Process: |
plasma membrane organization
[ISS]
phospholipid biosynthetic process [ISS] |
| Molecular Function: |
glycerol-3-phosphate O-acyltransferase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLELTPHSGQ KPFKKLFDWG VLFPYVAQVV RSEKFEYRQV TEIVHNDAVL KHAIKQAAEQ 60
61 TLREQRYAKN GHRLLTRSAS GDQEAQEEED EKRGISYQAI LRKQEQRAIS ILKDMGSTLN 120
121 NGLLAFTSWI LYKLLPCFLS GVVTNTKQIE MLKTATERSP GTPLIFVPLH RSHLDYIMVT 180
181 WILTNNDIRS PLVAAGNNLQ IPVFGGLLRG LGAFFIKRKI DPVEGKKDVL YRAALHLYLT 240
241 HALKQGHNVE FFIEGGRTRT GKPCMPKGGI LSVIVNAFMD GSIPDALLVP VSVNYERLVD 300
301 GNFVREQKGE KKIPESFGKA ISGIWKALKS NYGLMRIDFN EPYSIRELVN SYNKIAREDG 360
361 NIAKVYKPSA RVLQHNQSTS SLYGTDVVCE EHRNLIESIS RQVVFDCAAA TSVMSTNALA 420
421 FLLLTRFRNG AEEQILSEAL DDLRNSLSGC KDIGFSGESS QIVAYACDLL GSGLVTRSRD 480
481 ENGRLVIKAV NSVESFIELA YYSNMLTPHF ALSSILLTTF HSLLPETENK KEAAVSRKKL 540
541 IDTALENCQI YRYEFILNKP TQVLENLLYQ QLDDLLISGC VLTEKLHDDL PNGAEGRRLA 600
601 NVLAECLDED GYEDVRDGEA DEPKLLFASE TPSQQRYICE VLAPFAWTYV TVAQSLQILH 660
661 KNSMLESEFI SFVINDLSSK VKRGSCIYAE SISTDSVRNC LKLLEKWSVI EVCNQQGMRL 720
721 ISLNTLYEMS RESLNSIVKK IDAIVPKFNS GYFSVTP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
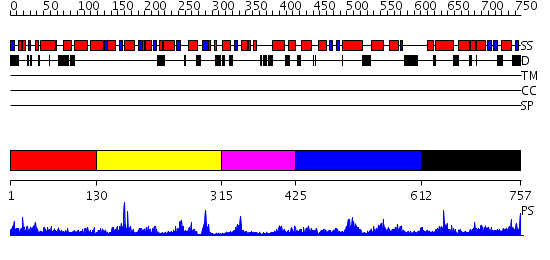
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 3.718996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [130..314] | 9.154902 | The 1.55 A Crystal Structure of Glycerol-3-Phosphate Acyltransferase | |
| 3 | View Details | [315..424] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [425..611] | 1.178981 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [612..757] | 1.094977 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)