













| General Information: |
|
| Name(s) found: |
CG5987-PA /
FBpp0084550
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein modification process
[IEA]
|
| Molecular Function: |
tubulin-tyrosine ligase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNLKNSKFFK RIQELRHFSK EKIRKDREKR THVVRALDMA LNEQPCLDRE WFSPEQSPPL 60
61 QPMSPRRRMK KTRCLKRRTA ADKQVDDNSP YYGPSSGENS PMPAEEAKLQ GFIQLNNKTY 120
121 RVECPTRVLN PPMNVDEERP ASETKSTICV SNSRYAMIGK ISKTLGYKLV KESKMWNILW 180
181 SDSFPGVELF KNMKRFQQIN HFPGMIEICR KDLLSRNLNR MLKIFPQDYK IFPKTWMLPA 240
241 DYGDAMNYAL NHKRTFILKP DSGAQGRGIW LTNDLKTIGP HERLICQTYI HRPLLIDGYK 300
301 FDLRVYTLIT SVDPLRIFVY NEGLARFATN KYVEPTPGNA NDLYMHLTNY SVNKRNSHYE 360
361 LCDNDDCGSK RKLSAINNWM RRHNYDVEEF WSNVDDVIIK TVLSAWPVLK HNYHACFPGH 420
421 DKIQACFEIL GFDILVDWKL KPYILEVNHS PSFHTNEQVD REVKRPLIRD TLNLVSTVLA 480
481 DKRQILKEDR KRVKQRLLKI RGDPPVQRPR LGGGTSKPKI DAQKGSPDAQ PVDVEPEAED 540
541 HGPLAQQIAW EESHLGNFRK IMPPPDLSKL DYYTRFYAQS HQVSIFAETA ASKKREDLAR 600
601 KMRIQLEEKK AKQQQMLNGK AKRDARKRHT VLLPRAVREK NRINFFRLRE NWSPGFISDA 660
661 EERLRHTWLH MRAEAIRTLK ITENIYSNLY ETGLLTNTDM VVYPHLYHNL QHGYDIKRTP 720
721 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
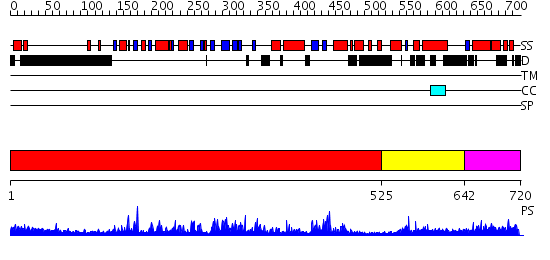
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..524] | 1.33 | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | |
| 2 | View Details | [525..641] | 4.166992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [642..720] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)