













| General Information: |
|
| Name(s) found: |
CG5871-PA /
FBpp0083452
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1019 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
hyalurononglucosaminidase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADEAGSQAD GKRQFICGVI EGFYGRPWTT EQRKDLFRKL KSMGMGSSPS YMYAPKDDYK 60
61 HRAYWRELYT VEEADHLSSL IAAAKEAGIT FYYALSPGLD MTYSSPKEIA TLKRKLDQVA 120
121 QFGCEAYALL FDDIESELSK ADKEVFQTFA NAHVSVTNEI YTHLGSPRFL FCPTQYCASR 180
181 AVPTVQESEY LNTLGSKLNN EIDILWTGDK VISKNISLES IQEITEVLRR PPCIWDNLHA 240
241 NDYDQKRIFM GPYSGRSPEL IPHLRGVMTN PNCEFYGNFV AIHSLAFWSR CSLDSKVNSS 300
301 LSADIKLETE NDDDLPAEFL SKNVYHPRLA LKNAITEWLP EFFMKKEAWG PITKPQPQVQ 360
361 MVMPIIPIIP SINTCMSLTT TTTTSTSSRT VPPTVNTTQL QALADVCVVT SSLTPISNPV 420
421 MNSLVSPTKV ITNDDIINPI PTTAASNIEL PKKIPISVVP VPIMETKSVE ASVELALDNA 480
481 VFDDNEIEPN SDSVKERLEL EVNLEGKQEP VANLSVDTML DDDSLSPLSG VVNEPMECSS 540
541 SITSQVSPRE EEAIKVVADD VLMESVNDVH SMHVESGTSS PISNAEMREE TEAQSDRTND 600
601 NNTIEGEGIT VDDLVLLCDL FYLPFEHGSR GHKLLVEFNW LKGNANVILQ DRSAGGGGDA 660
661 IKSDKPEVSE WHQRREQFDQ LCSAVVELLI KIANCPNKEI CHELYSYMWD ISGALSLLNC 720
721 YVKWLALGHF PQNTSSYTEG SYTWFSKGWK EAFMSGDQEP WVFRGGLIAD LQRLMPVDSG 780
781 NDLFVYKLPE QPTANYYLLR PYCNSDEQQV NDLCTRLYLQ WRGELDGGRH IPFPLPANVP 840
841 NIVADGLIGG YLTLSPQLCI VAYDESNRII GYSCAALDVN IFRRNLELCW YTELREKYSR 900
901 DICPLEGGEE VVQLVTSLVE SYHDSSGNGA LDQCPVEVSG SFPAVLISGT LREAEERDSG 960
961 ITKRMLTVLL AALRANGCFG AHVRVPQQDV AQVNFYSRIG FVDVYREEAT KCIYMGRRF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
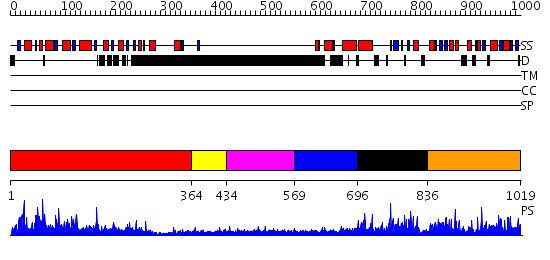
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..363] | 97.0 | No description for 2chnA was found. | |
| 2 | View Details | [364..433] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [434..568] | 4.305988 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [569..695] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [696..835] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [836..1019] | 4.09691 | Hypothetical protein YqiY |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)