













| General Information: |
|
| Name(s) found: |
sec15-PA /
FBpp0083436
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 766 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA]
cytoplasmic vesicle [IDA] axon terminus [IDA] exocyst [ISS][NAS] |
| Biological Process: |
synaptic vesicle targeting
[ISS][NAS]
synaptic vesicle docking during exocytosis [ISS][NAS] vesicle-mediated transport [ISS] phototaxis [IMP] axon guidance [IMP] bristle development [IMP] neurotransmitter secretion [NAS] endocytic recycling [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKVTVQDIE AVDDYWGPTF RSILEGNNTK QIGDQLEQRI RSHDKEIERI CNLYYQGFID 60
61 SIQELLQVRT QAQQLHNEVH SLDTSLRQIS ASLIQQGNDL VRARQIESNL ASAIEALKSC 120
121 LPALECYMKF TQQAKNKQYY QALRTLETLE TEHLTRLKTH NYRFATQMQI QIPIIKENIR 180
181 RSSASDFREF LENIRKFSPR IGELAITHTK QLQKRDINAI IAEHMQQMNG GEAGGAGAGG 240
241 DDDGANVSAQ DLIDFSPIYR CLHIYMVLGQ REYFEKDYRQ QRRDQAKLVL QPPPNMHDNL 300
301 EAYKTYICAI VGFFVVEDHV KNTAGDVVTS SYLEDLWSSS LTKFVNEISM SSSSCTDPNI 360
361 LLRIKNLIML SINTFKCYGY TVNILWELLH NMRDHYNEVL LQRWVHVFRE ILDKEQFLPM 420
421 VVQNTEEYEC IIERFPFHSE QLENAPFPKK FPFSRMVPEV YHQAKEFMYA CMKFAEELTL 480
481 SPNEVAAMVR KAANLLLTRS FSGCLSVVFR QPSITLTQLI QIIIDTQYLE KAGPFLDEFV 540
541 CHMTNTERSV SQTPSAMFHV ARQDAEKQVG LRICSKIDEF FELSAYDWLL VEPPGIASAF 600
601 ITDMISYLKS TFDSFAFKLP HIAQAACRRT FEHIAEKIYS IMYDEDVKQI STGALTQINL 660
661 DLMQCEFFAA SEPVPGLKEG ELSKYFLRNR QLLDLLILEE WSTYFHDYGK QENRYHLVQP 720
721 QSIIVILEKI READKKPIFS LVRKNDKKKL LETVLKQLKH IADRQN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
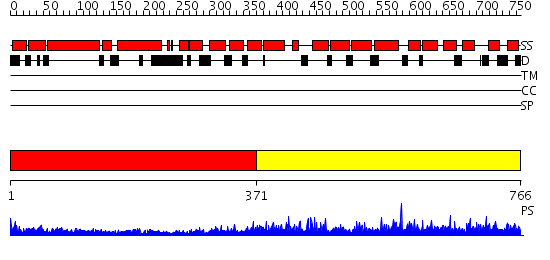
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..370] | 16.045757 | Heavy meromyosin subfragment | |
| 2 | View Details | [371..766] | 104.0 | Crystal Structure of Sec15 C-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)