













| General Information: |
|
| Name(s) found: |
l(3)07882-PA /
FBpp0082928
[FlyBase]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 852 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[ISS]
|
| Biological Process: |
maturation of SSU-rRNA
[ISS]
ribosomal small subunit biogenesis [ISS] rRNA processing [ISS] |
| Molecular Function: |
snoRNA binding
[ISS]
protein binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAKGKKASA DAVYAKKTTR SANPFDNSTA QSSKRGNPFD VHVNKEKFKI LGRICKHDRG 60
61 MPGVSRAKAL QKRAQTLGQQ FAVKHKTNRF KDNRIGKHLS GDQLTESVMN ARYLAEKMSQ 120
121 VRSNQKAEKF NLNDDELLTH RGQTLEEIEQ YRDERSDDEE LDDEALDADF TAAAHFGGEG 180
181 DSAQDRQAAI DEMIVEQKRR KNEIAKEKDE VYDLTEKLDA NYKLLLPLVA KVTKDEQDAK 240
241 PPPDAYDKLL KEMIFEPRGS VADKLINPDE LAKQEAARLE KLENERLRRM RADGEEDEEA 300
301 SVAKPKHRSA DDLDDGYFLA GEDDEGDDTL AYDLDGNLGT HLNGKKEAVL KGDENEDDDD 360
361 KEGEEEEEED SDEESDSEVD NLSDLKESES ESEPEEAPKA SKKKAKKSAD TIPASLDTSI 420
421 PFTIKMPKTY EDFTELLSKH ATAQKAIIIE RIIKCNHPKL EGVNRENVVK LYSFLLQYLK 480
481 DLFEDASEQD IREHFQLLSK LMPYLYELTQ LNPERMSNTL LEVIKEKYEE FRKNHKMYPS 540
541 LDTLVYFKLV ANLYSTSDFR HPVVTPCFIF IQHVLSRSRV RTRQEISMGL FLVTVVLEFV 600
601 SQSKRLVPAV FNFLQGIVHM SIPKRDVEQL EITPPFERDG PLSKLLALPA NTESTKLEPQ 660
661 QLQPTDLVTQ TITPDFKVRA LDTSLLLIKE ALQLVEEHVG ACYLAQPFLA LLSRLPLESY 720
721 PEHVHQHHKD ATELAEKLAA QKMKPLAPAE KKPKALRLLE PRFEAVYDDK RRPKMSKAKE 780
781 ERAKLLHKIK REKKGAIREI RRDTSFVQML RLKQTLQSDK ERHEKVKRIY QEASVQQGEL 840
841 NELSRAKKKK KF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
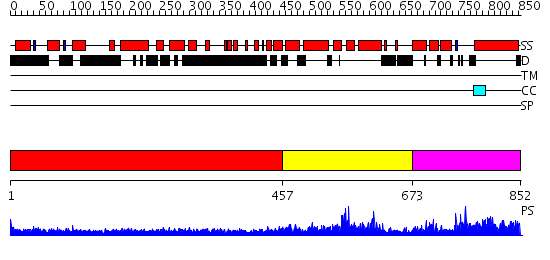
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..456] | 37.522879 | Heavy meromyosin subfragment | |
| 2 | View Details | [457..672] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [673..852] | 0.975 | No description for 2qagC was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)