













| General Information: |
|
| Name(s) found: |
Mical-PA /
FBpp0081684
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 810 amino acids |
Gene Ontology: |
|
| Cellular Component: |
terminal button
[IDA]
cytosol [NAS] plasma membrane [IDA] |
| Biological Process: |
sarcomere organization
[IMP]
axon guidance [IMP][TAS] neuron remodeling [IGI][IMP] dendrite morphogenesis [IGI][IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
actin binding [ISS] oxidoreductase activity [IEA] electron carrier activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRQHQRHHQ QHHHLPPHQQ PQQQMPQQQQ QLTAQQQQQQ QLLMAEHAAA AEAAELFDLL 60
61 CVATTMRQIL ALHRAMCEAV GLRPSPLNDF YPRLKAKVRS WKAQALWKKF DARAAHRVYG 120
121 KGAACTGTRV LVIGAGPCGL RTAIEAQLLG AKVVVLEKRD RITRNNVLHL WPFVITDLRN 180
181 LGAKKFYGKF CAGSIDHISI RQLQCMLLKV ALLLGVEIHE GVSFDHAVEP SGDGGGWRAA 240
241 VTPADHPVSH YEFDVLIGAD GKRNMLDFRR KEFRGKLAIA ITANFINKKT EAEAKVEEIS 300
301 GVAFIFNQAF FKELYGKTGI DLENIVYYKD ETHYFVMTAK KHSLIDKGVI IEDMADPGEL 360
361 LAPANVDTQK LHDYAREAAE FSTQYQMPNL EFAVNHYGKP DVAMFDFTSM FAAEMSCRVI 420
421 VRKGARLMQC LVGDSLLEPF WPTGSGCARG FLSSMDAAYA IKLWSNPQNS TLGVLAQRES 480
481 IYRLLNQTTP DTLQRDISAY TVDPATRYPN LNRESVNSWQ VKHLVDTDDP SILEQTFMDT 540
541 HALQTPHLDT PGRRKRRSGD LLPQGATLLR WISAQLHSYQ FIPELKEASD VFRNGRVLCA 600
601 LINRYRPDLI DYAATKDMSP VECNELSFAV LERELHIDRV MSAKQSLDLT ELESRIWLNY 660
661 LDQICDLFRG EIPHIKHPKM DFSDLRQKYR INHTHAQPDF SKLLATKPKA KSPMQDAVDI 720
721 PTTVQRRSVL EEERAKRQRR HEQLLNIGGG AAGAAAGVAG SGTGTTTQGQ NDTPRRSKKR 780
781 RQVDKTANIV SKCWHYFISW IGSYANVQKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
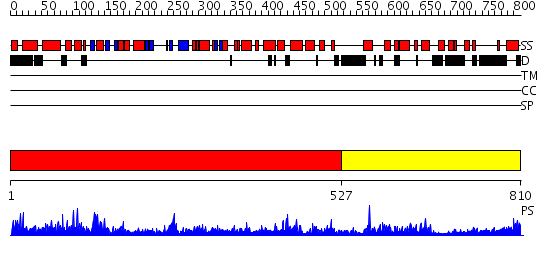
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..526] | 151.0 | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | |
| 2 | View Details | [527..810] | 101.0 | Cryo-EM Structure of Chicken Gizzard Smooth Muscle alpha-Actinin |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)