













| General Information: |
|
| Name(s) found: |
mura-PC /
FBpp0081596
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1173 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
olfactory learning
[IMP]
learning or memory [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNSNATGGN NHGGSSRRPF TRNSSSVRSQ RGGGGGLGGR MVYSPHTHPH VQQQQQQQQQ 60
61 QQQQGQQRSS NNGNNNLWLG NSPWCNVNLN GGNSGNNNNN GGNNNNNNGG NISSNNNSNN 120
121 VNNKDANPNC DVQTSNSSGT CIYNNSKAHH HHNNSNNNGH TQSHIHTNHS LHQGHHHPHH 180
181 TQMHTPHYPH SSSPQQSNSY RQYPPHSYSP NSPHSNGHTN SNASNNPISQ RSNPQAHPNQ 240
241 NQNSNFYEMC TGSGGSHTYG SMSVSLVRLN PARLCLTLGP VTPPAQRVVS FGHRSHSHSN 300
301 SSSNTSSSDQ NQHNPPPRSQ NHHPNPHHSQ YQYQYQYQYR YQYHPPHQYH PLHSPQQQTN 360
361 RQSLPLTRTN SNSINATTFN STNPTTNSNP TDVVNVNSCT DIIPYGSSST SSSASSMLPQ 420
421 QQQHQINNAS APINSHSQGQ GPGNSPNLGH NHQRGYNNGN NNRGIGNGIV NGNTNGPPDY 480
481 MNYRRGVCPA QSRDYSGNGN GNNNGHGHNG RRYMSDNLNN PSSSMNGLIN SGHHQRNYND 540
541 RNLNNHFGNS EQNGGPDHRD RAEGSSYNFM RNGGGSGGGG YGRNGSHYQH MAYGNNNGAS 600
601 TSTGGPGLMG ELPSGSGLSG SSLSLNNGPG GLNSDSPSRK RRRISGRPPN GPQPQHRCPM 660
661 AHMQQGSPPL RRPRLRDVAT STQQQQQQYG HQVHHPQQQQ QQPPSNYHPM HHHQPQPHYV 720
721 PQQQVPPPHV QRSPWDLGSS GGSSGGATSS SSVGPILQQV QAPPQPPSHQ QAVGYPQAPP 780
781 QPASLMVDLN LNQVPVNLQL RPSEPFWASF CTYPIPAQAR LAPCHLHGVY TQPFPAAPPP 840
841 GLAAPPLQQQ HQPLIQAQQM ISQATLTAQQ QQRDVVAIAT ANLGPIEAPG AHGHPHAHPH 900
901 AHPHAHQLPP GIHITPLSGA AAAAATHHLH STAAAAAAVA AGAQITTAQQ ILFSSDRRTF 960
961 PPHRRIPRFW TANHGHRHVL PPQSLAAHQA PVQIQATSGI INPGFLLNFL AMFPLSPYNQ1020
1021 HDLSSGDTNE TENYEALLSL AERLGEAKPR GLTRNEIDQL PSYKFNPEVH NGDQSSCVVC1080
1081 MCDFELRQLL RVLPCSHEFH AKCVDKWLRS NRTCPICRGN ASDYFDGVDQ QQQSQATAGA1140
1141 AAALSGTSGG SAGVAGTSEA SAATANPQQS QAA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
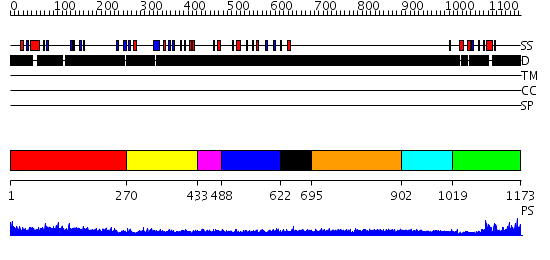
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..269] | 4.522879 | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | |
| 2 | View Details | [270..432] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [433..487] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [488..621] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [622..694] | 1.32 | Crystal structure of the C-terminal domain of BclA, the major antigen of the exosporium of the Bacillus anthracis spore. | |
| 6 | View Details | [695..901] | 5.221849 | Sec24 | |
| 7 | View Details | [902..1018] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1019..1173] | 9.221849 | Solution structure of RING finger in RING finger protein 38 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)