













| General Information: |
|
| Name(s) found: |
CG2926-PA /
FBpp0078401
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2296 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC]
|
| Biological Process: |
regulation of alternative nuclear mRNA splicing, via spliceosome
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDRDSDAVS DAGPRLSQQR GQRNRRIIMR IEFKDSEDSA SDQDRDDRPS CSAATRLRRS 60
61 TRRQEVASHS DSDSVPRKST RKLRNRYGPR SDCSSSGSDI DQLPTRTRKR KKVESSDEEE 120
121 PERTPNAVGR IQPMPADPDD GFSSDISSND LLEKCPICLL TFRQQEIGTP ATCEHIFCAA 180
181 CIDAWSRNVQ TCPIDRIEFD RIIVRDSYAS RRVVREVRLD LSKSNTELVV DDEADTAALS 240
241 EEEVTNCEIC ESPDREDVML LCDSCNQGYH MDCLDPPLYE IPAGSWYCDN CIDSDDEDDN 300
301 EQLELADDLD QLYEDIRGMG LPETRLRVRE VQEPPRILRT RQNERIRAAV LRRTRSGATS 360
361 AEDSTLTRTR TRTTTTTTTT TTTTRRTTTS TKKRPVQRRR RRRTRYRTYV VEYDLNNFDE 420
421 KFALKTSKKV IRRRRRRRRR GVATASSEGP GRRLSASKRL AEQMGVKSEG FQSSHLSGGT 480
481 ASSFSLFGNA NDLEYFSDSE PDAVSQISSA GLGSTAVQTS VRIGSIGAPR IRKALLQGKA 540
541 RIAAPPTAPT SAPAADILSS ILDLQDRWHD ASRNLGEVHI RADGTLNIPQ RPAPTATEAP 600
601 LPKAGTSKTE KHLTQAPLYQ QGGGGPNYNR GGGAGSSDSY NNRYSGNTSY RGGGGGGAGG 660
661 SSGGSGVPSS SGEASSRHLT GEGDGQGGGG FGNQNFSSSG SNNNTNMSSF IPFHLRFNTP 720
721 TRQQQPRQQN VLPANQPTPP FPGSAPPPAP VASSSNNFSG QTPMFAAPLN HHPAPVMGMP 780
781 VVLSIPPPPM PPASLPWNSP LFKITPLQQA PAKSNDGNQN DDVDNCPNFS IYSQESQAVA 840
841 NASAMPSGVP HGPADASDKD DDDMNEDLVQ LDDDDEDTDI PLPLGPEPEP VPEKVPKSSD 900
901 DDLYEPENPT EEPEEPEMFE ESCDAVPTEK SESSDHEPSN SNVQAAAPEE NDAEEARTRS 960
961 TPSPLVKEDR AADRDRAKGV LELYDDSDWE ELDIDKPKEF EKALLTETEA QVSPKRAPSK1020
1021 RPPSKKDTSL ERDVNESGSE HEQDRSYTPC LDEKNLAEEE EAGTTHQNNV STSPKTRSSK1080
1081 VSGDEDEPEK AVGTDIQSED ELNNENESPR RSRSRGTANN KKDKSKSRSK RNKGETFKKV1140
1141 GKRQKDRNYR GDKSEPDRSP RPDSRSLTPK MRSKSPRVSK SISRSRSGSR SRCKSKSFSR1200
1201 SRSRSPSRSR NRRRRNSRSK SYSRSHSRSR SRSNSHSRQR FSFRGRDHQR GFFRGRGGLR1260
1261 LNYRNQQFHN RPFQHYGQNY HQNQNYHHQH NQHQNYFPPN QHGQHQQYFQ RPKRRELPRY1320
1321 DVRNVVGTSR HQTQAKDRYG RDAMRSHRSR SGRRNSRSFS RSVSRDSRVS LRSRSGSPKR1380
1381 RSFSMSPTPP PGTSPPPHKH GRGRQTSTNR RYARSPSPAP SPRNERATSP VQSVHSRRSR1440
1441 SHTPNRINGD GNRSSLYMGS PHYTPRISRS RSRSRSKSPK DMHKKKKKKS DKKKRRKRTG1500
1501 SSSPTTAARM RRISRQRDPF EDADDFLAPQ RKRKKAGITT GQWSPSPSPG RCDFLHDSAL1560
1561 QDKNSSWTPP LGTPKGGHYN NDGGLTPKKT KRKRDKSKKK RKQHQLEKPR KEKKRKRRTQ1620
1621 TPEPLPSKEV FASGNNILVS VSFNKESNTN NASATLSSQQ QSVVTIPPGR EDLLSNRMTS1680
1681 MTSMASMASN SNICSAVGAV KKTKRKRKKL EAKPVAIIDL ERSPFQVHQE PADVIVLTDS1740
1741 EDATEPLSSR RERERDRDHE LMRENGQREK TPQVGSLDAI METSYENMTQ STGPKTPPEP1800
1801 PLVKFNLPTK KIQHKVRNNP LHEDVDDINS ADELEASCSM APAEQTPQHG HASHDGGQKI1860
1861 GPNTPPESGP CSPDAYDPFE PTKSPSLSPR SPTLTPSQNL DQQTVSMPAS GAIGPGSGDK1920
1921 GESDGAHVTG SNASTNTQAQ GNALNPVDLV MALMNKPNQS GSIQQESEVS SAQYISSSSL1980
1981 SLSLGVGSST GGGGGGAGGG DNHDKTADGK TITVLSNVLL TSSSVVSSSQ HIPSISSPTP2040
2041 PSKKLTPLPK AGGSVASSSS GVGNIPTTSS GIGGTRNGSG LGNAGGSSNA LDDSFSMEIE2100
2101 SPYSPGSADY EDLFEPPIPL EASRRAKGGA TGGKVEIFDN LFGSSSPISN IRMTSRYNTA2160
2161 VGKKSNRTKA DRKSKVKGAR NPDFYDDVPN SATDLQNKDR LLRKLNRQER VVEEVKLVLK2220
2221 PYFNKKAITK DDYKDIMRRA VPKICHSRSG EINPHKIKNL IDAYVRKFRA KHKKLSLLNT2280
2281 GQVSSAVKCA AYLKKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
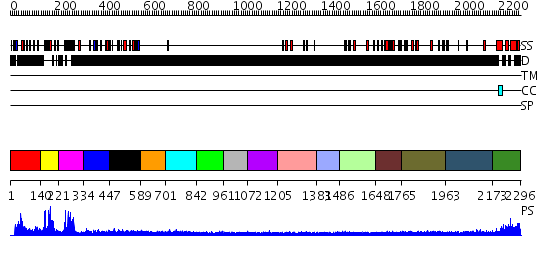
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [140..220] | 8.69897 | No description for 2ep4A was found. | |
| 3 | View Details | [221..333] | 17.30103 | No description for 2e6rA was found. | |
| 4 | View Details | [334..446] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [447..588] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [589..700] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [701..841] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [842..960] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [961..1071] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1072..1204] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1205..1382] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1383..1485] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1486..1647] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1648..1764] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [1765..1962] | N/A | No confident structure predictions are available. | |
| 16 | View Details | [1963..2172] | N/A | No confident structure predictions are available. | |
| 17 | View Details | [2173..2296] | 2.347993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)