













| General Information: |
|
| Name(s) found: |
Snr1-PA /
FBpp0078331
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 370 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS][NAS]
nuclear chromosome [IEA] brahma complex [IDA][NAS][TAS] |
| Biological Process: |
muscle organ development
[IMP]
chromatin remodeling [IEA] embryonic development via the syncytial blastoderm [NAS] positive regulation of transcription, DNA-dependent [IDA] negative regulation of cell proliferation [IMP] epidermis development [IMP] imaginal disc-derived wing vein specification [IMP] regulation of transcription [ISS][NAS] instar larval or pupal development [NAS] dendrite morphogenesis [IMP] |
| Molecular Function: |
RNA polymerase II transcription factor activity
[NAS]
transcription coactivator activity [IC] general RNA polymerase II transcription factor activity [ISS] SET domain binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALQTYGDKP VAFQLEEGGE YYYVGSEVGN YMRHFRGILY KKYPGMTRIV LSNEERKRLA 60
61 ESGLSSHILA SSVSLLRAVE VDDIMAGNDE KYRAVSVNTS DTPVPRESKS KKQPQYVPTM 120
121 PNSSHLDAVP QATPINRNRV HTKKVRTFPM CFDDTDPTAS LENAAQKECL VPIRLDMELE 180
181 GQKLRDTFTW NKNESMITPE QFAEVLCDDL DLNPLPFVPA IAQAIRQQIE AFPNDPPILE 240
241 ETCDQRVIVK LNIHVGNTSL VDQVEWDMSE KNNNPEEFAI KLCAELGLGG EFVTAIAYSI 300
301 RGQLSWHCRT YAFSEAPLST IDVPFRNPSD ADAWAPFLET LTDAEMEKKI RDQDRNTRRM 360
361 RRLANTTTGW |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
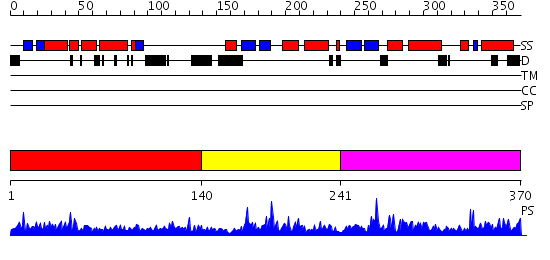
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | 1.019979 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [140..240] | 2.038988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [241..370] | 6.038979 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)