













| General Information: |
|
| Name(s) found: |
gi|7296815
[NCBI NR]
gi|24644079 [NCBI NR] |
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 934 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
protein modification process
[IEA]
|
| Molecular Function: |
ligand-dependent nuclear receptor binding
[ISS]
zinc ion binding [IEA] acid-amino acid ligase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAESVKSQSL SALTEGQHDD GGSTATSATL ANKTSGTNTT SASNHSSQRR RNNNTNSNRN 60
61 KSHHKNEKRN APATSGPAVS SSVDVVPATS TASATSTRRS RSQGRHSSAA ALSSKESQIP 120
121 KRSVGAIRSP SSPAFNSIPV VSDQSLSPGS RKRQLNQHNN SNNKASSSHQ SAPGGDQLVC 180
181 APLKKRRLQQ SLGSGICEGA VEINLGAASF DHTPRQASGD CVAQRTRSKT ISPEELPSTS 240
241 SAAAARHHQI RPSASSTSFS SSHRNKRKAS IGGSIAAVET TPHRGTAAAR AGRSGNLLNY 300
301 YRKTRKVSHT RSSQKSEKQS IPAEDTTANR NGSAGSGSSS KSGVIPKNRK SLRHSQQNLL 360
361 DTEAAEVESA AAETGGEQQS EHGNLDVIDQ LPTPEAGPNQ SEASGIHNQS QLEADRQPVV 420
421 QDEDDEEDDE EEEEEEEEEV GFYGIVNSAG SSYEEDTQIV AEEDEITTEE EVDDEDDDEI 480
481 EEEDLSESEF AQQLIGELGG ERQQPQRATN NAKKTKNNTP AVAAASTIKA TTTATTSSGY 540
541 QQRAAPHGQG AAPGAGGGGP RATRSSNSNS SNNVVDYSIM PHLTTVATTT PHALTPQQHQ 600
601 QQRQPPQPPS HQQQQVHQQQ QPPQLLPTHQ FAHLSSFVTP TAAAAAAAAA AHYPPASAAA 660
661 AYFAQQQHHQ HQHQHQQQHV QQAQAPQPHY YHSSVAVLHQ PPPHHFTSTG AGPPPALFQQ 720
721 QAPPQLTRYT PATATTAATF VPPQQQQVVY NPQQQQHSSL RRSSRGKTGS CVSSAAAAQQ 780
781 QQHHQQQQHS SAAAAVQQQL LPPPGTYQYQ QVGGNTVVVA VRHQQQLQQQ QQQQQQLALH 840
841 HQHQQQQHQQ QQQQHQQQQH QQQQRATLVQ PGFLFSYRSN QPQQQQQPAQ QIHQGPKVTH 900
901 SSGKKMKEEE EQNKDERTRD RKQIKIQGLL NHFP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
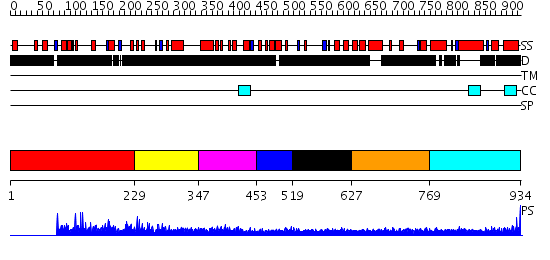
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..228] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [229..346] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [347..452] | 3.152996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [453..518] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [519..626] | 2.221995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [627..768] | 7.39794 | Sec24 | |
| 7 | View Details | [769..934] | 1.56 | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)