













| General Information: |
|
| Name(s) found: |
CG14650-PA /
FBpp0078566
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 970 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to heat
[ISS]
protein folding [IEA] |
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATRQDDEIG GGSTESAPFR VLPHTDEVGN YLAHQQQQQQ QQQHQHQTQN YLIHQQQHGQ 60
61 APVPLMWPPP PPHLAGPPFA PHLQQQQPHM YNEYLFNLSY SGQPGQPETY SVLPVGPGNF 120
121 LKVYHCPDNQ VSEAGTPLFT HINMASINHH QLQHQHQAAP PTPSQPTPLY ELFATNPLTL 180
181 PHAQQQDAQH RQPQQTQTQA PTVSQVNHSS SSANLLINNL VNNWSPNLTG GSYIQFDGEA 240
241 NEMDVVLETS IAQQPQVPNQ NLQQPEPLSQ SVNTLPAASK PAKSLKMPLA TATVSPTAIN 300
301 KSSVAVPGQP EGKKRIVAEV KPMPMSYSDV LSKGTKPSAA GRDMRADNGG DYSVQSQRRP 360
361 EKEDAYGNRE MRTSKRSPLH DAKETGSFVP GVGHAHASRG KKRSKVSQTA PLKVQQSQSL 420
421 AQPNLQPQIK SNKLNNPEKK RPLQPKNVNS NAPKFSEHKT TPVSANSSLQ NHSNVLNFTP 480
481 ATNSSTSSRK SGSNKINSNA SHSNHTGANT HAGSSSSSSS VNYSSNNNVS SSSAKRYSSS 540
541 NLNPNNSSGY SYSSKRNRSN AYSSTNSPTH ANSFSSNRNY ELAKRIIHTW WIYTLKLLTW 600
601 LFYLVYDIVV LGFSMGYERI TLAYVAALAY ARQLQKELKQ NSGKPSIWWR SYWRRFDARF 660
661 AKNSRLAVWR LFYKRKPPET TSEQFKTGRL PQTGEEAMYS LLNCKGKDAY SILGVPPDSS 720
721 QEQIRKHYKK IAVLVHPDKN KQAGAEEAFK VLQRAFELIG EPENRLIYDQ SIAETLHTEK 780
781 AWTELHDLLS QLQTKMAEAA NTIRCSTCAQ RHPRKLTERP HYAARECASC KIRHSAKDGD 840
841 IWAETSMMGL RWKYLALMDG KVYDITEWAN CQKGALSHLE PNSHMVQYRI VRGAQQQQQQ 900
901 QQQQQQQQQQ QHHQHPQQPH HDRGVHHPGG GVSGVSEATL HEFLDNLYSG QHPGAHNAFA 960
961 GNARRRTRRN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
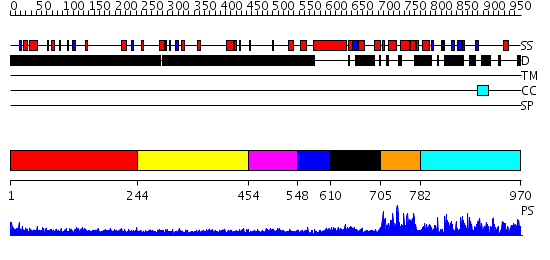
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..243] | 3.244994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [244..453] | 1.242984 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [454..547] | 3.190986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [548..609] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [610..704] | 5.0 | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | |
| 6 | View Details | [705..781] | 41.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 7 | View Details | [782..970] | 40.154902 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 | No functions predicted. | ||||||||||||
| 5 |
|
||||||||||||
| 6 |
|
||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)