













| General Information: |
|
| Name(s) found: |
CG6613-PA /
FBpp0071672
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
intracellular signaling pathway
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] diacylglycerol binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLFRSLNH FSSKRENVVK EALINTLNEN AREIEFEVKQ KSLEVLGVCE QTSALCTTLE 60
61 ALFLHGLKDS FLSATFNVIA GDVERRPEPS FWAPCMVFMH KQVIEQVQGL SQITSETGQC 120
121 RAWVRQSLNE SVFSSYLNNM RKNGSALSQY YKRNALMRDS EGLETAAKIM ESLEAYVQFD 180
181 LPVNSSLLNH WPDYSLQLSD LWTPALKSCP ISSGVDVASS LGSDIIAIPT PQPLQTNELF 240
241 SESISNSPFG RNSNFGVPEL SQRENLDLLI QKFDEMDVIN EEQTGGDKAE TNETAEQEAP 300
301 SGSKTRSRKK SNRQMDSSSF ADLDLEAPSL LPQGSNSLSN LMQTSWSGDL EATPTSPTEE 360
361 LTGSRFFQRS VSVSSSVSLR SPTTDRCSYN ALLRKHESNR ESGAGWSEIW EKFRASASNL 420
421 NVAQPQRDTD PPISEDLSDL QSLDTNSEFE LLTSEFDMVE LQQMVAQLCQ LAREPGLDAQ 480
481 GFLCKSCQHP LGIGYSNFQV CAFSGSYYCN SCMDVEMQLI PARIIYNWDF RKYSVSKRAA 540
541 TFLAEFRSHP FLDMQLLNPR IYFASDAMAE LQSLRIRLNF IRAYLYTCAP SSIELLQNQF 600
601 AGREYLYEHI HLYSIADLAL IQRGVLCQQL QKAFKLGEAH VLKCRLCHLK GFICEICQSP 660
661 RVLYPFHIST TFRCLACGAV FHAECLNEKQ PCPKCERIRK REDQGLQEAV QCLKQKNEVA 720
721 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
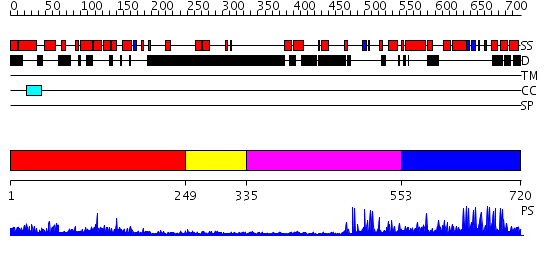
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | 45.09691 | Crystal structure of the RUN domain of mouse Rap2 interacting protein x | |
| 2 | View Details | [249..334] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [335..552] | 8.522879 | Hrs | |
| 4 | View Details | [553..720] | 1.26 | Crystal Structure of The Cand1-Cul1-Roc1 Complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)