













| General Information: |
|
| Name(s) found: |
FBpp0301119
[FlyBase]
FBpp0301116 [FlyBase] FBpp0301112 [FlyBase] |
| Description(s) found:
Found 41 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1156 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA]
germline ring canal [IDA][NAS][TAS] female germline ring canal inner rim [TAS] fusome [IDA][NAS][TAS] spectrosome [IDA][TAS] plasma membrane [IDA] |
| Biological Process: |
ovarian fusome organization
[IMP]
cystoblast division [IMP] testicular fusome organization [IMP] centrosome organization [IMP] germarium-derived oocyte fate determination [TAS] female germ-line cyst formation [TAS] germ-line cyst formation [TAS] female germline ring canal formation, actin assembly [IMP][NAS][TAS] fusome organization [TAS] spectrosome organization [TAS] meiotic spindle organization [IMP] |
| Molecular Function: |
actin binding
[ISS]
metal ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEVEQPPQN GIDPTAGEDD DNSKARPADI EQDMREMERR KRVEAIMGSK LFREELERIV 60
61 DSARDGGAGA SGILQQLSDI VGVPVSRVGS VFKSSNCMVP INDIRGVESM GYAKGEKILR 120
121 CKLAATFRLL DLYGWTQGLG AQITARLKVD QEYFLVNPYG LLYHEITASA LNKVDMQGQI 180
181 VEQGTTNFGG NKSHFVLHSV VHAARPDIRC AIYIGCSPVV AISSLKTGLL PLTKDACVLG 240
241 EITTHAYTGL FDEEERNRLV RSLGPNSKVI LLTNHGALCC GETIEEAFFA ACHIVQACET 300
301 QLKLLPVGLD NLVLIPEESR KAIYEQSRRP PEDLEKKFAA VAAAEDGAAT AEKDAAEAVP 360
361 KVGSPPKWRV GGAEFEALMR MLDNAGYRTG YIYRHPLIKS DPPKPKNDVE LPPAVSSLGY 420
421 LLEEEELFRQ GIWKKGDIRK GGDRSRWLNS PNVYQKVEVL ETGTPDPKKI TKWVAEGSPT 480
481 HSTPVRIEDP LQFVPAGTNP REFKRVQQLI KDNRRADKIS AGPQSHILEG VTWDEASRLK 540
541 DATVSQAGDH VVMMGAASKG IIQRGFQHNA TVYKAPYAKN PFDNVTDDEL NEYKRTVERK 600
601 KKSVHGEYTD TDFSESEAVL QAGTKKYPQS EPETEHQVIE IQTQQAPVPR QAEVVLSDAL 660
661 VSQLAQKYAF LYSPGQYMYA CMKMAPLMHK VYVIHKVEPV SKHNYPPVND GNMSIHHNES 720
721 GAGFMAQESS VISSTPVRNA LASVSLPEER NHSILGLSST PYRTISHFGF NCPLITSPTI 780
781 LLHPEHRSIW QRVAEQREKV VSFIDLTTLS LDNRKLLNVV TSTHPTQCQS QSQSFISEKH 840
841 IQLEVTPPKR KQRVYSATIS SGLDDSLDEL DSLMSGLAIN MPRSREQDSG LYRSYTFLPS 900
901 NHALPKDTDA NNRDQTDRER PEAEQEESFH CAGDSGIGDS TGRRPRLATT SNDSSIQEAE 960
961 AYTQGKHVKL TLSSSPTPTA TQSPATIEIL INVSLRNAEC VQTVQTHEQE FRAKLERVID1020
1021 EEIHYISQQL AFKQRQAELH EQQTTSRAPI ATPSFTTMHP PAPASSSSMV HRSNSAPELC1080
1081 HTYSYVAVGD LSTKQDQASP QLPAEGEPLN DILSSLEKEL ERLLNSVVTA HMLHNKAIIH1140
1141 ECRARFSQLA DGIVSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
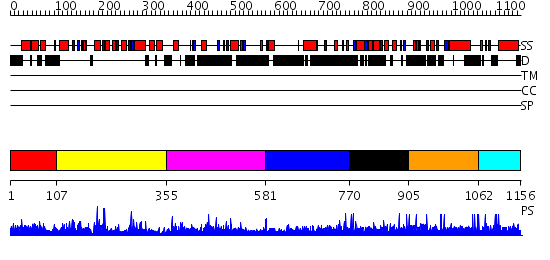
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 1.108973 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [107..354] | 54.0 | L-ribulose-5-phosphate 4-epimerase | |
| 3 | View Details | [355..580] | 1.308965 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [581..769] | 7.324997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [770..904] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [905..1061] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1062..1156] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)