













| General Information: |
|
| Name(s) found: |
CG8613-PA /
FBpp0086674
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 808 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
mannose metabolic process
[IEA]
|
| Molecular Function: |
mannose-6-phosphate isomerase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRVRPLLPR IRSSLQTTLK LQADTSQSES ESFRRNLRLL HNSCRSRPVS NQKFRTMATG 60
61 GEASKEVSKE EPAKQGNRLV ASKSPYLLQH AYNPVDWYPW GEEAFEKARS ENKIIFLSVG 120
121 YSTCHWCHVM EHESFENPET AAIMNENFVN IKVDREERPD IDKIYMQFLL MSKGSGGWPM 180
181 SVWLTPTLAP LVAGTYFPPK SRYGMPSFNT VLKSIARKWE TDKESLLATG SSLLSALQKN 240
241 QDASAVPEAA FGAGSAIEKL SEAINVHRQR FDQTHGGFGS EPKFPEVPRL NFLFHGYLVT 300
301 KDPDVLDMVI ETLTQIGKGG IHDHIFGGFA RYATTQDWHN VHFEKMLYDQ GQLMMAFANA 360
361 YKVTRDEIYL RYADKIHKYL IKDLRHPLGG FYAGEDADSL PTHEDKVKVE GAFYAWTWDE 420
421 IQAAFKDQAQ RFDDITPERA FEIYAYHYGL KPPGNVPAYS DPHGHLTGKN ILIVRGSEED 480
481 TCANFKLEED RFKKLLATTN DILHVIRDKR PRPHLDTKII CAWNGLVLSG LCKLGNCYSA 540
541 NREQYMQTAK ELLDFLRKEM YDPEQKLLIR SCYGVAVGDE TLEKNASQID GFLDDYAFLI 600
601 KGLLDYYKAT LDVDVLHWAK ALQDTQDKLF WDERNGAYFF SQQDAPNVIV RLKEDHDGAE 660
661 PCGNSVSAHN LVLLAHYYDE NAYLQKAGKL LNFFADVSPF GHALPEMLSA LLMHENGLDL 720
721 VAVVGPDSPD TQRFVEICRK FFIPSMIIVH VDPSNPEEAS NQRLQTKFKM VGGKTTVYIC 780
781 HERACRMPVT DPQQLEDNLM AYFFSKRD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
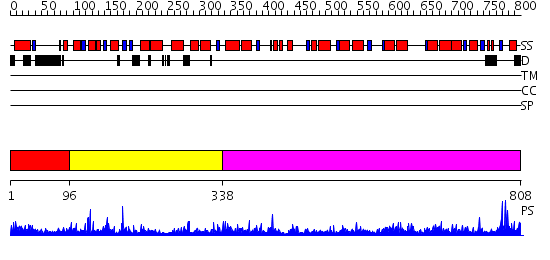
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [96..337] | 16.69897 | Crystal Structure of Yeast Protein Disulfide Isomerase | |
| 3 | View Details | [338..808] | 57.69897 | N-acyl-D-glucosamine 2-epimerase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)