













| General Information: |
|
| Name(s) found: |
FBpp0308787
[FlyBase]
CG17019-PA / FBpp0086919 [FlyBase] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
negative regulation of apoptosis
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPCESCGVEF TVFRRKRACF DCKRYYCANC IASRRCKRCS VFAQRPLSRT DLLKLKPKDL 60
61 IFYLQSKHIS TEGCLEKEEL VGLVLTHVAQ VDRSRGSNAS SNSPGRGQAN PIDSLKQSCQ 120
121 NFFSNLSDNI SDSLASFDSK VTTKPPESRQ PEVPTHIFEQ PRVSTREIPT YASAVNGGPQ 180
181 HVHVQQQATT TNGGSQSSSS PSTSTQSSNN RDASSSSQAH VAHQEARNEE QEDAQSKSDC 240
241 ECSDEEIMAT FSERVSISLK TDTSGRRVNA AAEPSPVDLA NVDNGPAGNT APSPGCSKPG 300
301 SSSSKADASS QSSFEELGAI GGISDESKAN TDTNSSHLDQ WQVLEVNRIE AVEETPTTSA 360
361 ADEILEVTLR SRPAVEGDDS EMGESTQALN VNQRPANPSP AFYPAVPQRT KKVTRRRSDG 420
421 YLNRRYHSSD DESPVAPGGP GLSLGLSTLS EHPEHGLHPH SHRQSCQRCG KNKTNIRRHV 480
481 EKMRRHLENS QMSEEDIKRE LQEFLTYLEQ RTKSVDASDA DSHAVSPLSV DAISVAAGGR 540
541 SPDMPITPTI EFSPTQDEDI RWDDDEGIHV YAAPSDFEPT AGIDSRFVNL EDFEDLKDLE 600
601 GLTVKQLKEV LMLHRVDYKG CCEKQELLDR VSRLWKTMRE CPAVEKLATD ELCKICMDAP 660
661 IECVFLECGH MATCTSCGKV LNECPICRQY IVRVVRFFRA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
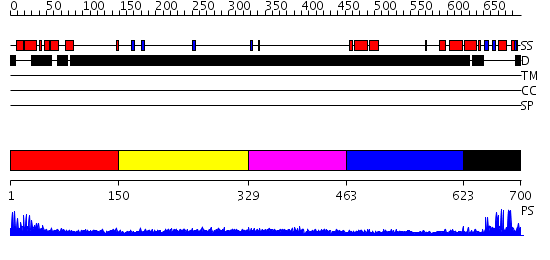
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 20.154902 | Crystal Structure of a FYVE-type domain from caspase regulator CARP2 | |
| 2 | View Details | [150..328] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [329..462] | 4.174981 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [463..622] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [623..700] | 13.522879 | Cbl; N-terminal domain of cbl (N-cbl); CBL |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)