













| General Information: |
|
| Name(s) found: |
ana3-PA /
FBpp0087015
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1977 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
centrosome duplication
[IMP]
centriole replication [IMP] mitotic spindle organization [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTKQSPALQ LAEGQLTKLT SESEEIRMRA LDQIETRFIR CLQLGEPIQF KPVLLLKQLI 60
61 RWFGYTPPLV PDRVLAMIME LLRSEYAEAV IRKIPYERFK AELQKVRRVL HKQESKRVSE 120
121 LLDDMNLLLL EKYNIDRVTP SVSSLSSNDI PSQATESADS SSNQIYDNLK PEDYEPSWSH 180
181 PCLDDVATMK SMIDLPRNSV ELQLQLTELI IRMGDYPTEY FLQPPFVFLH LVQLQTMTDG 240
241 SLIHVNRALI ACLRLLQQRI LLRRNTLSYA DRFDPPSRPK QVKVVSALVI LLENCMKLIR 300
301 PLLFSCTNDN WHIMELIVEI VRTHDVLSSK IPLVSITLIA DAVKSLLAYC NSVEGSCMTQ 360
361 LMDSLRIPRL QSLILNGVLH DMVALNINYD KRMDRRQAKA LIQPIVLDSA YLSGMPERMK 420
421 SLNTLISSLD SGPSPDEELI KLKRAYSVAL NQLHPNTELT GSLLIQKYRQ VCLVLVQLGS 480
481 ETLVKQLFGA VVKCIPFYAG NLKLRKDADE LLYTLVDLPA LKLRSLIYRL MIKSTVAHFH 540
541 SFMNKTVYMT GCSNVDLIRQ HILGVPLTPL LLRRMIIQSS EANTSERMQQ WCLDYPIMIM 600
601 KLNAILAPQD FSVVFPLLLP VLPLLISRSV THKILHNVIW NVLEPDSSRL DPQLMLRGYV 660
661 YFMFHPDGEV RSEATTTIAY VLQCQEQTNR YLPTASNVPV EHIANDLCII QPPFCYRSIF 720
721 IKCSDERFQG QRSLDALFRL LQAKDIKSNI RKSTMTQLNV LLRNWRACEE FSTKEDGYRL 780
781 IMESLHNALK KGNDTDILLP TVSILMKLLF NDDEFRLEVA NTFGVYTLLL RALFLFPHLE 840
841 QLRQDVSICL FQMLFQNCIT STEDKLVLNA NMEPLILPVT YEIEHKVIPT AVTEGLELQQ 900
901 NVEATHFGRD RAKAAQHWRL YMAYRVCQVP SSITLESVSA LDIRESLKIK MADLALVRSS 960
961 DLNEQLSCQF IAAENCSKHE DLQKTVSAIQ LFLVVLRNSL SDTVAENLWK LIHKYIRLAP1020
1021 GNEADDKVYK SMLDLCVTCI RFSQPHAING LSYALETDHH HSFQLLLHDS QISLDKLFLI1080
1081 CQCMMQLLSN ELSDDSMNWY GKFFMQLSAV AKTHFELRQL QHVRCILCML RFVSERNLKF1140
1141 SNAQLMSYSQ HFIQLSSNLR TSTQTGSEWQ RDCLYIICQL QIHLIYQEPK ASSKASIPND1200
1201 VGASYKVLRY FLTLCGHGDS EVRALSWVSL ANWITICGSQ VAEILPRLDF LPGGLPACCL1260
1261 TTLTDAHEMM LIRELAGRVF ILLMPLIGAE SSLELLRKHD LLKTAYNSLK AIHDTPWMFK1320
1321 KIVGERHSCE VISCYVAICT KMVAMKPEWC AALCGHSLMS GLSDVMKKLK SQASCSIPLV1380
1381 ELCASQICEL YSMCYTNNFE FLKMTICRDS VFLQNYLTLI NDVLSLECPE YMVIPLLKMF1440
1441 LIFCTDSNSN SFLIEQIKNK PSLFMDFFLF GLHVKLINSP FQRFTLSALS LVFTKAQLAA1500
1501 YDISMLSELE KFELAFSDPD IAKDGKQEFE SKKKQSVSQC IYDESSDNSD DNKQRPNHAI1560
1561 QATNAAIIIF HRLDQLFDRY YLSKALNFLE LPAVGQVQVC EALGELLKAS TWAVKAAGES1620
1621 KLLGKVVHIL DDFLNDEKIG NAAVYVKRVG PHKAQSIISN LLVLINMLSQ WHSSPNSEII1680
1681 QTSMAANIAK ILIRIWPWLS HSYHLKKVTV QLIMFLTEHS FEMCKQISLL QSGHAQSLLH1740
1741 LMARVADFET TKKEIPNKEP SLNMVPALRV MVNCCCCTEG RLSLSKMNLL DMFDTILPAN1800
1801 PCSTHLKVRP PVLIAWLGFW EVFSRYDVGG KACHLQSLIN TIRRSPPLSH KRILCLRILR1860
1861 NMCFFNGNRP RLVELADFIN LLRDILEQRV QKAPTSEKNG LNSFEEHRLA VLMLWKLFGF1920
1921 GAKYKGMLRG TKLFKLLIGL RVEMSVVFSE EENKYAGVPY ANDLAKLLEK LMESMRQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
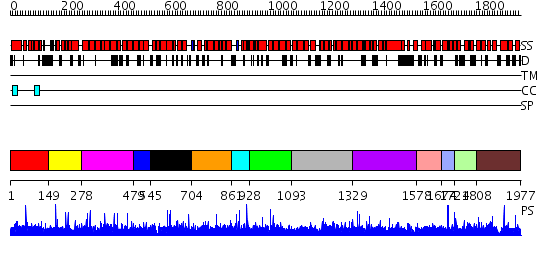
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [149..277] | 1.007988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [278..478] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [479..544] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [545..703] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [704..860] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [861..927] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [928..1092] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1093..1328] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1329..1577] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1578..1673] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1674..1723] | 1.007992 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [1724..1807] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1808..1977] | 1.010958 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)