













| General Information: |
|
| Name(s) found: |
FBpp0300187
[FlyBase]
FBpp0300186 [FlyBase] FBpp0300185 [FlyBase] vlc-PF / FBpp0289730 [FlyBase] vlc-PE / FBpp0289729 [FlyBase] vlc-PC / FBpp0085377 [FlyBase] vlc-PA / FBpp0085375 [FlyBase] |
| Description(s) found:
Found 72 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 605 amino acids |
Gene Ontology: |
|
| Cellular Component: |
septate junction
[ISS]
|
| Biological Process: |
imaginal disc-derived leg morphogenesis
[NAS]
cell-cell signaling [IEA] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDRSLDSIGS CSLDVDADSS DISDTSGSLN FPTPVSVRDI TREFTANLRE RCVPNLPPQP 60
61 QHKTPAYVES RSSELEHMVL SPAIVECDQE PSSNDFDETS AENSQEAPEK KPSYLNLACC 120
121 VNGYSNLTTY DSKIRQDINK SREVSPIRPS TSSLQYCKRN HFLATPTLHS MPSPATKCKP 180
181 IEPNNNSNDS SGNFSSGSGH SNGNGSFIKQ RVERLYGPGA LAQGFYSPKK HGQSPSNAAG 240
241 RSERGQVEID SLQGSELARK FKQLSPSKDY GEFRKKMQKK CSQSTTARID EPLHSPGLNG 300
301 DLDLPVFRHL SQEFRAQLPT VSPKRGAPRS CMAPDVLLVD NESSPDHGPT DEVDHESTKM 360
361 DKLASEEYIS TSNISVEAVV KDGNFFLKIL KEEQSRLLVL AALAEKYADA LSTNPDISED 420
421 TFGLLRSASG KARLLVSQKM KQFEGLCHNN LNRSPEDKFP TTLDDLQGFW DMVYLQVEHV 480
481 DSIFADIEKL KSNDWKVKAQ SVADQPDLHT IPTKTIKPSK VGVSQVKTVG KASSVRVNGS 540
541 APPPPNSAAA LKREAQRKLL LEMKRQRREA MTAAAKLNPI NVDADPDVPG IIGDSCANPS 600
601 VSNGS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
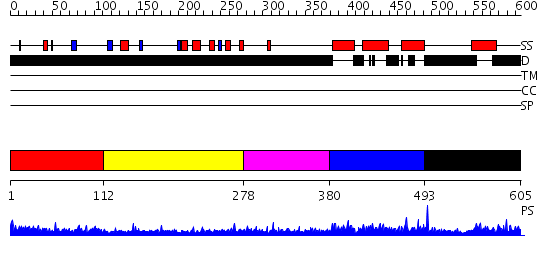
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [112..277] | 7.139996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [278..379] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [380..492] | 2.122975 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [493..605] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)