













| General Information: |
|
| Name(s) found: |
gi|116310433
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Indica Group |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEVADLENPT SRSSSQKSSR RSGSRRSQKS AGQQSSPTVF PEKKGKSKSS RQKHLVFDNK 60
61 DSKKAKNNEQ KNDVVDEKSN FSGYEIYSGK LFFDKKNRIT GDQISANGKA DISDVRLTSK 120
121 ALIWGSSLLS LEDVISVSYS SGVQHFIVHA YPSKKYLFGK THRVRKDLRF IAPTVEEAIS 180
181 WVTCFAEQNI YVNMLPLPPT SSTEQDLDGP LSGALFDHPP IKCRTPPRIL VILNPRSGHG 240
241 RSCKVFHDKA EPIFKLAGFH MEVVKTTHAG HAKSLASTFD FSAFPDGIVC VGGDGIVNEV 300
301 FNGLLSRSDR AEAVSIPVGI IPAGSDNSLV WTVLGVKDPI SASLLIVKGG FTALDILSVE 360
361 WIQSGLIHFG TTVSYYGFIS DVLELSEKYQ KKFGPLRYFV AGILKFFCLP SYFYELEYLP 420
421 SSKEMTGHGK GIGQENFVSD VYTNVMRSRS KREGIPRASS LSSIDSIMTP SRMSLGDVDT 480
481 SGSTAASTEP SEYVRGLDPK AKRLSLGRSN IVSEPEEVLH PQPHHSSFWP RTRSKTRTER 540
541 NSVGVTTNDT RLSWAAPSIH DKEDISSTIS DPGPIWDSEP KWDNGPKWDT ELTWESDHPI 600
601 ELPGPPEDNE IGPSVELVPN LDDKWVVRKG HFLGVLVCNH SCKTVQSLSS QVIAPKANHD 660
661 DNTLDLLLVG GKGRWKLLKF FILLQFGRHL SLPYVEYVKV KSVKLKPGAN THNGCGIDGE 720
721 LCRVKGHVLC SLLQEQCMLI GRQSRQST |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
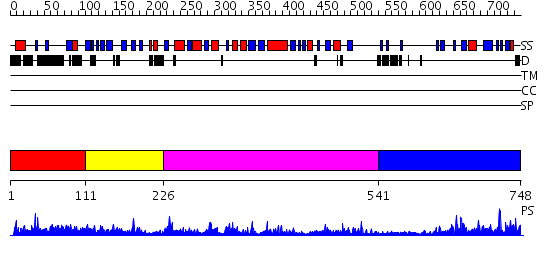
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [111..225] | 12.0 | Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta | |
| 3 | View Details | [226..540] | 37.69897 | No description for 2jgrA was found. | |
| 4 | View Details | [541..748] | 2.66 | No description for 2bonA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)