













| General Information: |
|
| Name(s) found: |
gi|124000261
[NCBI NR]
gi|123997205 [NCBI NR] gi|123993297 [NCBI NR] |
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 334 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKRRDILAI VLIVLPWTLL ITVWHQSTLA PLLAVHKDEG SDPRRETPPG ADPREYCTSD 60
61 RDIVEVVRTE YVYTRPPPWS DTLPTIHVVT PTYSRPVQKA ELTRMANTLL HVPNLHWLVV 120
121 EDAPRRTPLT ARLLRDTGLN YTHLHVETPR NYKLRGDARD PRIPRGTMQR NLALRWLRET 180
181 FPRNSSQPGV VYFADDDNTY SLELFEEMRS TRRVSVWPVA FVGGLRYEAP RVNGAGKVVG 240
241 WKTVFDPHRP FAIDMAGFAV NLRLILQRSQ AYFKLRGVKG GYQESSLLRE LVTLNDLEPK 300
301 AANCTKILVW HTRTEKPVLV NEGKKGFTDP SVEI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
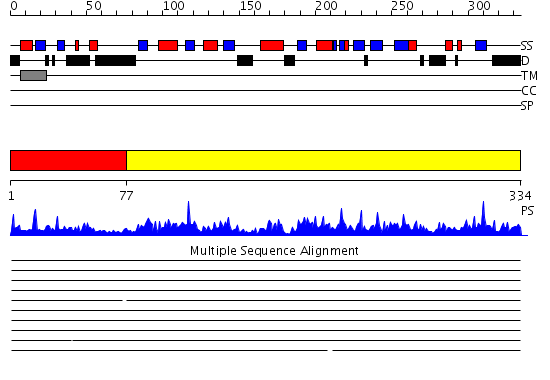
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [77..334] | 115.0 | Crystal structure of human GlcAT-P apo form |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|