













| General Information: |
|
| Name(s) found: |
gi|123999198
[NCBI NR]
gi|123988683 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 619 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADEGKSYSE HDDERVNFPQ RKKKGRGPFR WKYGEGNRRS GRGGSGIRSS RLEEDDGDVA 60
61 MSDAQDGPRV RYNPYTTRPN RRGDTWHDRD RIHVTVRRDR APPERGGAGT SQDGTSKNWF 120
121 KITIPYGRKY DKAWLLSMIQ SKCSVPFTPI EFHYENTRAQ FFVEDASTAS ALKAVNYKIL 180
181 DRENRRISII INSSAPPHTI LNELKPEQVE QLKLIMSKRY DGSQQALDLK GLRSDPDLVA 240
241 QNIDVVLNRR SCMAATLRII EENIPELLSL NLSNNRLYRL DDMSSIVQKA PNLKILNLSG 300
301 NELKSERELD KIKGLKLEEL WLDGNSLCDT FRDQSTYISA IRERFPKLLR LDGHELPPPI 360
361 AFDVEAPTTL PPCKGSYFGT ENLKSLVLHF LQQYYAIYDS GDRQGLLDAY HDGACCSLSI 420
421 PFIPQNPARS SLAEYFKDSR NVKKLKDPTL RFRLLKHTRL NVVAFLNELP KTQHDVNSFV 480
481 VDISAQTSTL LCFSVNGVFK EVDGKSRDSL RAFTRTFIAV PASNSGLCIV NDELFVRNAS 540
541 SEEIQRAFAM PAPTPSSSPV PTLSPEQQEM LQAFSTQSGM NLEWSQKCLQ DNNWDYTRSA 600
601 QAFTHLKAKG EIPEVAFMK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
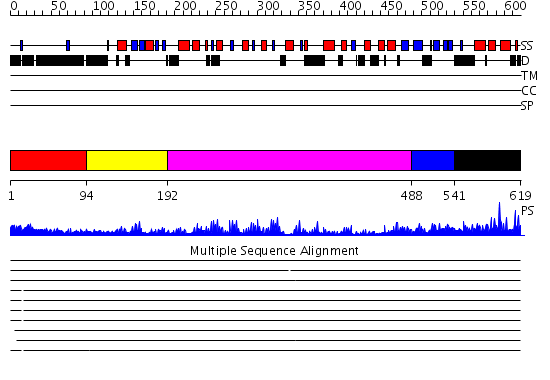
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [94..191] | 41.30103 | mRNA export factor tap | |
| 3 | View Details | [192..487] | 21.69897 | No description for 2id5A was found. | |
| 4 | View Details | [488..540] | 15.69897 | Crystal structure of the C. albicans Mtr2-Mex67 M domain complex | |
| 5 | View Details | [541..619] | 76.045757 | NTF2-like domain of Tip associating protein, TAP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)