













| General Information: |
|
| Name(s) found: |
ipla-1 /
CE40099
[WormBase]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 779 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
metal ion binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLGKKKVSR ATKSDDTQPD PNGKYTMAQT KDADLSRPSG LPSNGNPGSS TTVPPASKSG 60
61 PVAPPRPSGT PVAPQRNRRR KVTELKCSEV RWFFQEPKGT LWNPFNGRDS IMLEIKYRKE 120
121 KGIELDEAMQ EIYDESLTHY KMEMKDEPEI ENGNIGMEQE KPMVVVMNGQ YKVNKDNSKI 180
181 DPIYWKDDSK EIRRGSWFSP DYQPLEMPLS DQIEKNHLQC FRNQMIPEGT TVFSKSETSN 240
241 KPVLAELHVD GYDIRWSSVI DISLHQKGNA ILRYLWAKST PLRRGYEKEA DWNDAAAEIS 300
301 HLILVVHGIG QKGYENLIAQ NANQVRDGVV SAMEKVYPEE KSRPMFLPVE WRSALKLDNG 360
361 LTDNITIPKM SSMRASLNST AMDVMYYQSP LFRTEIVRGV VSQLNRTYKL FKANNPQFNG 420
421 HVSVFGHSLG SVICYDVLTQ YSPLMLFDKY VTKSIDEYLK RDDTNASEEA RKALEAMKLA 480
481 REQLRDNLEG GIHKLLVTKE EQLEFKVKYL FAVGSPLGVF LTMRGGESTD LLSKATNVER 540
541 VFNIFHPYDP VAYRLEPFFA PEYRHIRPIK LFSNTDLRAR ASYENLPLDV YKHYLKKLKN 600
601 LNKAKKNKDD KTADARSGGD DENEDEDECD SDEDARSGCS SPRSMTPPPF ETAAANAAAA 660
661 AKETKAVKKG WFSFGTSSNP KKTQSTASLG SVNATSTENI EFAKEAAEEL PLAEKILGSG 720
721 VRVPHRIDFQ LQPALTEKSY WSVLKSHFAY WTNADLALFL ANVLYCKPLK PEEAKPTWA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
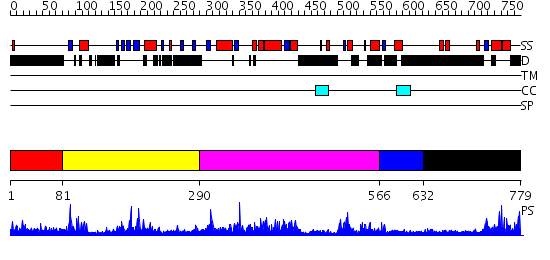
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..289] | 3.090978 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [290..565] | 1.03 | Lipase L1 | |
| 4 | View Details | [566..631] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [632..779] | 5.093984 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)