













| General Information: |
|
| Name(s) found: |
EFR3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 798 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWLLFRSKHK KLVLRCFPSG KLGETEPNGS PLAYLSYYAA SNSSKLRKVA HFLGSRVRHS 60
61 YYHKRDNEVI IGLKICKTLV QKCRDNINVM ASEVVNMLLV ASSSKNLEVL SACVDCFATF 120
121 CDNSGKGSPA TFGNEFHSAF NNLVNSFFEL SKGIDCVDPQ QSKMLGLKAF HALTACKFAG 180
181 TEGGMRYQPH FAIHCVLINS WSESDQEKKS FIDLVRSTAK SYKPLPPSST GVSLPEHDEA 240
241 GDNIDVAIKK LCIRILFNIY TQTDPLFMAE STKSLIHFFA AKSDTPVNLE YISVVLNQIL 300
301 DWTPVELRHS IFFCCLRCLS SSRIVNSETN VMVPYMIYSI LNSKNSISGL SVIDVLRDLG 360
361 SHLINAVNSF DADDQTWYNM CESPSEKLPR RLYLLLVCIT CLTKHQYYDE EFADVWREID 420
421 TLANSETSSA IQMVALTAFH KLQNEKLNTK KESSAPLSLV TDFLLHSWPK SAERLHTSQS 480
481 PFVRIKTAEV FYEFLCFLRP SLINFDTILR KSAITSFSVL WDFIYETSWH IERWLKVFSV 540
541 QSEFYILKLI IQRLYQLYGP VSVTAVLPAM YRGLRPFEND PPRCIAEAVT AYYIIYIGEN 600
601 LKITTLQSSG RKWLDSLSSS LPSSLLNRGS STHSWEKLTN ASTEEVMLPL DMVVNELLEK 660
661 SPKVFPEDSR LLFAEQSRHP KYTDIIQKLK KPSREKSFTS SSEYSLPFIS PASDYQQNPL 720
721 LHATKSLVSI HQQSQRGEMV STLKQALSRP HTASTVVRSP SEINLTRQTS NRVPLLDMLN 780
781 LNRAMSPTPI QSPPYVRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
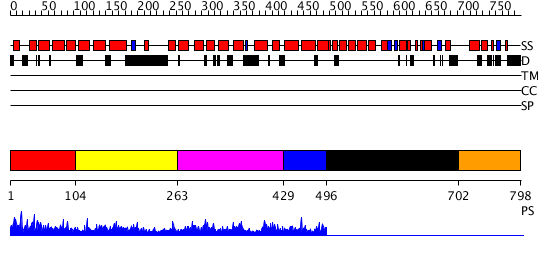
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | 2.047978 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [104..262] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [263..428] | 1.047983 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [429..495] | 3.036996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [496..701] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [702..798] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)