













| General Information: |
|
| Name(s) found: |
SOG2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 886 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAHEVSSTK NSEIERIIKE AEDAGPENAL TLDLSHLNLR ELPYEQLERI QGRIARLALG 60
61 HNFIKSIGPE ILKFTRLRYL NIRSNVLREF PESLCRLESL EILDISRNKI KQLPESFGAL 120
121 MNLKVLSISK NRLFELPTYI AHMPNLEILK IENNHIVFPP PHIANNDLQD SDMQLFIANI 180
181 KGYLSRNESV FRTRSESTVS AALSASANLS HSEENDSSVV DSYLFSAPSD TSHAVSPGML 240
241 TFVTPSPHSH SPAGHQQSTP KSTLSKTNEN SEGTLYDSNV AHGCTHPPSL NQLNKFHLDA 300
301 SPRQARPRRS VSLATGLNSP SVSKPPSSAT GPLYHSPQSS LTNSSVASAD VQERTHNTNG 360
361 ASPIQDQISE FTDQHQNPSN NDAASTQSIL KTAPTQLSAS AKTSAISLPE VAKKERNRSN 420
421 STNDDYSSTR LPSSVLHRLE ALKEARKLSE QLPNRIFAQD PHPSPRLKKE ETHENGSNLT 480
481 NDSVNSYFSN IGGSEVEMKH SAFEKTLESS RGILFSLSQV QQALRQQLLF CSNPVVLDSM 540
541 RHVLHTANVQ IKRLILCFED TQQSNDGTAN INSIVNASLS CISSFRKLIE VTKKFLNELT 600
601 SRADVRYVRL LLLILFDAAK ELQNALVPLS PSQPHSGNNI FADQVRQSPT SMIRTASGLT 660
661 NRIVSVEKPA SVMNPEIDKQ IQDQVALATN SAMSVLTLLI DAVPKNNQPD LVNENIAPNL 720
721 ISKLRELGSL SAAACDVTRK LKHRLLTFQT NPEELEWQLF LDDTQAFVKS IIAVANLAKA 780
781 LSSEYTFQKS VLAGLSAATR STKDLTILLS SSARHYTESS LATPVPLMSP IARVPATPLS 840
841 AALGSAAQSI TSPLIMSPAA IPASAYSTNK IDYFTDADGN VEGLQR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
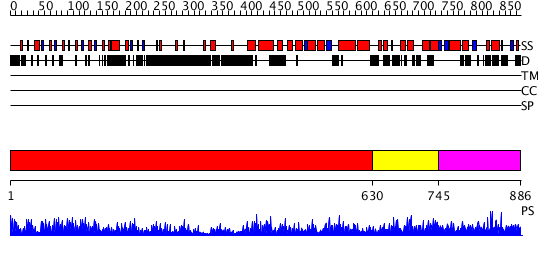
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..629] | 40.09691 | The molecular structure of toll-like receptor 3 ligand binding domain | |
| 2 | View Details | [630..744] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [745..886] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)